Team Finds 'Weakest Link' in the Aging Proteome
Proteins are the chief actors in cells, carrying out the duties specified by information encoded in our genes. Most proteins live only two days or less, ensuring that those damaged by inevitable chemical modifications are replaced with new functional copies.
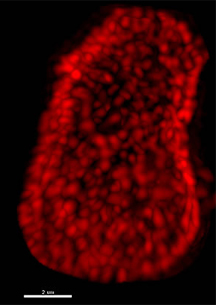
In the cover story of the August 29, 2013 issue of the journal Cell, a team led by researchers at The Scripps Research Institute (TSRI) and the Salk Institute for Biological Studies have now identified a small subset of proteins in the brain that persist for longer, even more than a year, without being replaced. These long-lived proteins have lifespans significantly longer than the typical protein, and their identification may be relevant to understanding the molecular basis of aging.
"Protein longevity can be a major contributor to cellular aging," said Martin Hetzer, a professor in Salk's Molecular and Cell Biology Laboratory and holder of the Jesse and Caryl Philips Foundation Chair, who was a senior author of the study with TSRI Professor John Yates. "Simply identifying all long-lived proteins allows us to focus our studies on these specific proteins, which may be the weakest link in the aging proteome."
“While a few extremely long-lived proteins with lifetimes of months or years were previously known, this work now shows that many more extremely long-lived proteins exist than previously thought,” said Jeffrey Savas, a TSRI postdoctoral fellow in Yates’s lab who was a co-first author of the study with Brandon Toyama, a postdoctoral fellow in Hetzer's laboratory.
System-Wide Identification
The study provides the first comprehensive and unbiased identification of the long-lived proteome, the entire set of proteins expressed by a genome under a given set of environmental conditions. In a study published in Science last year, Yates, Hetzer and their colleagues identified long-lived proteins in one sub-cellular location, namely the nucleus.
The new study takes the Science findings one step further by providing a system-wide identification of proteins with long lifespans in the rat brain, a laboratory model of human biology. The scientists found that long-lived proteins included those involved in gene expression, neuronal cell communication and enzymatic processes, as well as members of the nuclear pore complex (NPC), which is responsible for all traffic into and out of the nucleus.
Furthermore, the scientists found that the NPC undergoes slow but finite turnover through the exchange of smaller sub-complexes, not whole NPCs, which may help clear inevitable accumulation of damaged components.
"This can be thought of as similar to maintaining a car, where you don't replace the entire car, just the components that have broken down," explained Toyama.
Paving the Way to Future Insights
Savas points out the long-term collaboration between TSRI and Salk brings together biological insight, biochemical knowhow, proteomic mastery and bioinformatic innovation, paving the way for future insights.
“Future work includes the search for additional long-lived proteins in other tissues and those resistant to traditional degradation pathways,” Savas said, “as well as exploring potential links to neurodegenerative disease.”
Toyoma adds, "Now that we have identified these long-lived proteins, we can begin to examine how they may be affected in aging and what the cell does to compensate for inevitable damage. We're starting to think about how to get functionality back to that younger version of the protein."
In addition to Yates, Herzer, Savas and Toyama, other researchers on the study, “Identification of Long-Lived Proteins Reveals Exceptional Stability of Essential Cellular Structures,” were Sung Kyu Robin Park of TSRI, who provided bioinformatics expertise, and Michael S. Harris and Nicholas T. Ingolia of the Carnegie Institute for Science. For more information, see http://www.cell.com/abstract/S0092-8674(13)00945-8#Summary
The work was supported by the National Institutes of Health (R01GM098749, F32AG039127, RR011823, AG031097, MH067880), the Hewitt Foundation, the Glenn Foundation for Medical Research, the American Cancer Society (P30CA014195), the Ellison Medical Foundation and the Searle Scholars Program.
Send comments to: press[at]scripps.edu