TSRI Researchers Explore Ways That a Drug Like Avandia Can Be Made Safer
With the heightened concerns over the dangerous side effects of the once-popular antidiabetic drug Avandia, researchers at The Scripps Research Institute (TSRI) in Jupiter, Florida, are working to understand how small molecules, like those in Avandia, can have such varied effects throughout the body. The insights could help researchers design new drugs with better efficacy and fewer side effects.
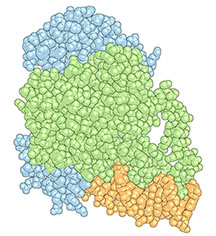
Douglas Kojetin, an associate professor at TSRI, and his team recently published a study in the journal Structure, showing the ways that Avandia interacts with and changes the shape of a combination of proteins, receptors and DNA—called the “complex”—resulting in the drug’s effects. In addition to helping inform the design of future antidiabetic drugs, the study revealed that DNA plays an active role in determining the structure of the complex, a finding that has implications for understanding how any small molecule drug affects the body.
Researchers in Kojetin’s lab focus on nuclear receptors—proteins that can interact or “bind” to both genetic material, such as DNA and small molecules, as well as bind to other proteins called coregulatory proteins that impact gene expression. Avandia, for example, binds to PPARγ, a nuclear receptor, which binds to DNA sequences important in regulating fat storage and metabolism. But PPARγ does not work with Avandia alone. A second nuclear receptor, RXRα, interacts with PPARγ to form a “heterodimer”, a complex made up of two different proteins bound together that recruits a co-regulatory protein called Steroid Receptor Coactivator-2 (SRC-2) to influence the activity of many different genes. As each piece binds, the complex changes shape, like pieces of wet clay mashing together to form a bowl.
The team set out to understand how specific sections of DNA affect the interaction between the nuclear receptor and coregulatory protein. For example, would this nuclear receptor interact with the coregulatory protein the same way if it weren’t bound to DNA?
Adding an extra layer of complexity, SRC-2 is an intrinsically disordered protein—a “floppy” protein without a consistent secondary structure that flops around like a loose spaghetti noodle. This means that popular methods that researchers would typically use to understand the protein’s structure, like x-ray crystallography, which requires a stable unflopy sample, wouldn’t be able to tell the scientists very much about what this protein is doing.
Kojetin’s team used a combination of quantitative biochemical, biophysical and solution structural methods to form a detailed understanding of these molecular interactions. Each technique gave his team pieces of information that they used to build a picture of how all of these molecules interact.
“No one tool could have given us the answer. It was really the combination of all these tools that gave us a full picture of what was going on,” Kojetin said.
It turned out that what was going on was a “thermodynamic” mechanism by which binding to DNA caused the receptor heterodimer to change its shape and importantly stabilize its floppy regions.
Kojetin’s team showed that DNA interaction impacted the potency of Avandia and its ability to recruit the coregulatory protein. The receptors on their own are like two balloons randomly floating around in the wind, explained Kojetin. When it bound to the DNA, the receptors become stabilized, as if the balloons were tied down together, making it easier for SRC-2 to interact with it.
If the proteins had bound to another portion of DNA, the complex may have been able to form a different shape and exert a different, possibly even dangerous effect, or perhaps no effect at all. Different genes are exposed, or accessible, in different bodily tissues. This could help explain how drugs can have one effect in a certain area of the body and a different effect in other areas where the sections of accessible DNA are different.
“The drug does not control activity in the body by itself. When the receptor is bound to different DNA sequences, the activity of the drug could be changed.” Knowing this, said Kojetin, “is going to open up a lot of possibilities.”
In addition to Kojetin, other authors of the study titled, “Synergistic Regulation of Coregulator/Nuclear Receptor Interaction by Ligand and DNA,” include first author Ian de Vera, as well as Jie Zheng, Scott Novick, Jinsai Shang, Travis Hughes, Richard Brust, Paola Munoz-Tello, William Gardner, Jr., David Marciano, Xiangming Kong and Patrick Griffin – all of whom work at the Scripps Florida campus.
The study was supported in part by the James and Esther King Biomedical Research Program, Florida Department of Health (1KN-09); the William R. Kenan, Jr. Charitable Trust (TSRI High School Student Summer Internship Program); National Institutes of Health (NIH) grants DK101871 (DJK), DK105825 (PRG), F32DK097890 (TH), K99DK103116 (TH) and F32DK108442 (RB); and American Heart Association (AHA) fellowship awards 12POST12050025 (TH) and 16POST27780018 (RB).Send comments to: press[at]scripps.edu