Coding the brain to decode disease
By Madeline McCurry-Schmidt
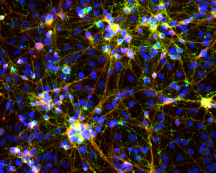
Scientists at The Scripps Research Institute have devised what they call a “neuronal cookbook” for turning skin cells into different types of neurons. As reported in the journal Nature, the research opens the door to studying common brain conditions such as autism, schizophrenia, addiction and Alzheimer’s disease under reproducible conditions in a dish.
“The brain is incredibly complex, with thousands of different types of cells that are each involved in different diseases,” says Kristin Baldwin, PhD, professor at Scripps Research and senior author of the study. “The problem with understanding and treating the many disorders of the brain is that we cannot reproducibly produce the right types of brain cells. Now we have found more than 75 new ways to rapidly and reproducibly turn skin cells into neurons that we think will be much better representatives of different neurologic diseases than were previously available.
“Having a personalized and nearly unlimited supply of different types of neuronal cells in a dish lets you uncover what’s going wrong in a disease. At the same time, the study supplies a new toolkit to test thousands of drugs on the affected cells to try to reverse the problems, rather than having to test them in mice or other animals, with results that are often difficult to interpret for human conditions,” Baldwin adds.
The Baldwin lab wondered if it would be possible to both simplify and expand the coding toolbox for making neuronal cells directly from skin cells. First author Rachel Tsunemoto, PhD, had hints from a previous study that it might be possible to generate very specific types of neurons using only two transcription factors at a time. So, she and other lab members engineered and tested a large set of two factor codes to see if they could convert skin cells into cells with the essential core traits of neurons, such as their shape and electrical excitability.
While they had expected to find a handful of new factors, or possibly none at all, results of their large screen were quite surprising. Of the almost 600 factors tested, over 12 percent ended up producing neurons—leading to more than 70 new recipes or codes for neuronal production.
Next came an even bigger surprise. The “synthetic neurons,” as Baldwin calls them, started to grow synapses and try to communicate with each other. “This came just two to three weeks after they were fibroblast cells that would normally never communicate,” says Baldwin.
“This was really great to see,” says Tsunemoto, co-first author of the study and a researcher with Scripps Research and the University of California, San Diego at the time of the study. “But obviously, there was a lot more follow-up work to be done to understand this exciting neuronal complexity in a dish in more detail.”
For years, the challenge has been to see past the traits that neurons share—like the ability to communicate using connections called synapses—and figure out why certain neurons have special properties, like the ability to produce dopamine or respond to neuroactive drugs such as nicotine, that also correlate with their involvement in different diseases.
Together with Sohyon Lee, a co-first author of the study and a recent PhD graduate at Scripps Research, Tsunemoto began to sort through the “outputs” of the different codes using traditional electrical recording methods and new sensitive sequencing methods to see whether the codes would produce neurons with different features.
What they found was exciting. Each code produced a set of neurons with different properties, some of which seemed immediately useful for understanding how differences in our genetic makeups can predispose us to neurologic diversity in disorders such as autism, nicotine addiction or neurodegeneration.
Baldwin notes that this work builds on years of research from her lab and around the world. Previous studies led by Nobel laureate Shinya Yamanaka and Marius Wernig’s group at Stanford University recently showed that sets of three to four factors could convert skin cells into pluripotent stem cells and directly into neurons. Previous work from Baldwin’s lab showed that sets of just two factors could selectively generate specific kinds of neurons that respond to pain and itch.
The new study is a big step forward in cellular reprogramming. The transcription factors to make neurons are like codes,” Baldwin says, and now researchers can input these codes to get the precise neuronal types they need for research, over and over again, eliminating the need for very precious human samples obtained from brain surgeries (which can be studied for only a few hours).
“Now we can be better genome detectives,” says Baldwin. “Building up a database of these codes and the types of neurons they produce can help us directly link genomic studies of human brain disease to a molecular understanding of what goes wrong with neurons, which is the key to finding and targeting treatments.
“We’re already working with collaborators and writing grants to apply this platform to autism and Alzheimer’s disease research, as well as to some rare diseases, such as Friedrich’s ataxia,” adds Baldwin. “We also think the transcription factor pairs we studied here are only the tip of the iceberg and there are likely many more codes for neuronal types that can be found.”
Researchers are encouraging other scientists to use their results through a freely available platform called BioGPS, run by Scripps Research scientists. “There’s so much to explore,” says Tsunemoto.
In addition to Baldwin, Tsunemoto and Lee, authors of the study, “Diverse reprogramming codes for neuronal identity,” were Attila Szűcs of the University of California, San Diego and Eötvös Loránd University; and Pavel Chubukov, Irina Sokolova, Joel W. Blanchard, Kevin T. Eade, Jacob Bruggemann, Chunlei Wu, Ali Torkamani and Pietro Paolo Sanna of Scripps Research.
The study was supported by the National Institutes of Health (grants DP1AG055944, DA031566, 5 UL1 TR001114, U54GM114833, DC012592, MH102698), the Dorris Neuroscience Center, the California Institute for Regenerative Medicine, a National Science Foundation Predoctoral Fellowship and the Andrea Elizabeth Vogt Memorial Award and the National Brain Research Program of Hungary (grant KTIA_NAP_13-2014-0018).
Send comments to: press[at]scripps.edu