Microscopic chariots deliver molecules within our cells
By Madeline McCurry-Schmidt
On the cellular highway, motor proteins called dyneins rule the road. Dyneins “walk” along structures called microtubules to deliver cellular cargo, such as signaling molecules and organelles, to different parts of a cell. Without dynein on the job, cells cannot divide and people can develop neurological diseases.
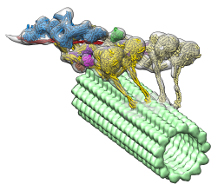
Now a new study, which appears in the journal Nature Structural & Molecular Biology, provides the first three-dimensional (3D) visualization of the dynein-dynactin complex bound to microtubules. The study leaders from The Scripps Research Institute (TSRI) report that a protein called dynactin hitches two dyneins together, like a yoke locking together a pair of draft horses.
“If you want a team of horses to move in one direction, you need to line them up,” says Gabriel C. Lander, PhD, a TSRI associate professor and senior author of the study. “That’s exactly what dynactin is doing to dynein molecules.”
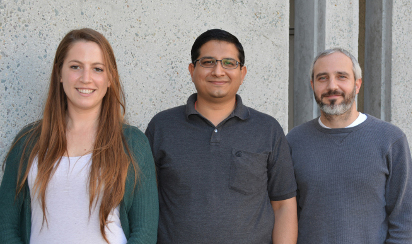
The study was led at TSRI by (left to right) Danielle Grotjahn, Saikat Chowdhury, PhD, and Gabriel C. Lander, PhD. (Photo by Madeline McCurry-Schmidt)
Understanding how the dynein-dynactin complex is assembled and organized provides a critical foundation to explain the underlying causes of several dynein-related neurodegenerative diseases such as spinal muscular atrophy (SMA) and Charcot-Marie-Tooth (CMT) disease.
Researchers knew that dynactin is required for dynein to move cargo, but they struggled to get a complete picture of how the different parts of the complex worked together.
“We knew that dynein only becomes active when it binds with a partner called dynactin. The problem was that, historically, it was difficult to solve this structure because it is very flexible and dynamic,” explains Danielle Grotjahn, a TSRI graduate student and co-first author of the study. “We needed to visualize these dynein-dynactin complexes to fundamentally understand how it works to transport molecules.”
For the new study, researchers used an imaging technique called cryo-electron tomography, which is similar to acquiring a CT scan of a protein, to produce a 3D reconstruction, or tomogram. To increase the resolution and clarity of these tomograms, researchers used advanced computational algorithms to average together many individual protein complexes.
In this case, however, the current processing methods were unable to produce a clear, interpretable 3D structure of the extremely flexible dynein-dynactin complex.
Grotjahn knew the necessary information was hidden in the tomograms, so she developed a new strategy to identify specific components of the dynein-dynactin complex. This approach helped guide the image processing software to reconstruct the entire microtubule-bound motor complex.
“Danielle was doing the first big step for the computational algorithms,” says Lander.
With their new, detailed structure, the researchers noticed a surprising feature: the complex has two dynein molecules where they expected to only see one. This means because each dynein has two motor domains, the dynein-dynactin complex has four motor domains total.
“This discovery was totally unexpected, and will change how this motor complex is represented in cell biology and biochemistry text books,” says Saikat Chowdhury, PhD, a TSRI research associate and co-first author of the study.
“There had been years of biophysical experiments and biochemical experiments, and it was always assumed that there was just one dynein molecule,” Lander adds.
The researchers could also see where dynactin fit in. They found that the molecule provides a stable scaffold, hitching together the team of motor domains and activating their ability to move along microtubules. This discovery helps explain how dynein can haul large loads, such as organelles that are much bigger than themselves, over long distances in a crowded cellular environment.
The image processing approach used in this study has the potential to be extremely useful for solving the structures of other large, flexible proteins.
“We’re now able to move past cartoon models and visualize the fine details of many dynamic macromolecular complexes,” says Grotjahn. “As we learn more about the 3D organization and architecture of these molecular machines, we will be better equipped to understand how they malfunction in disease.”
Additional authors of the study, “Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility,” were Yiru Xu of The Scripps Research Institute; Richard J. McKenney of the University of California, Davis; and Trina A. Schroer of Johns Hopkins University.
The study was supported by the National Institutes of Health (grant DP2EB020402, R00NS089428 and S10OD021634), the National Science Foundation, The Searle Scholars Program, the Pew Scholars Program and the Johns Hopkins Krieger School of Arts and Sciences.
Send comments to: press[at]scripps.edu