Crouching Protein, Hidden Enzyme
By Madeline McCurry-Schmidt
Meet a microscopic gymnast.
A new study led by scientists at The Scripps Research Institute (TSRI) and the University of California (UC), Berkeley shows how a crucial molecular enzyme starts in a tucked-in somersault position and flips out when it encounters the right target.
The new findings, published recently in the journal eLife, give scientists a clearer picture of the process through which cells eliminate proteins that promote diseases such as cancer and Alzheimer’s.
“Having an atomic-resolution structure and a better understanding of this mechanism gives us the ability to someday design therapeutics to combat cancer and neurodegeneration,” said TSRI biologist Gabriel Lander, who was co-senior of author of the study with Andreas Martin of UC Berkeley.
Keeping Cells Healthy
The new study sheds light on the proteasome, a molecular machine that serves as a recycling center in cells. Proteasomes break down spent or damaged proteins and can even eliminate harmful misfolded proteins observed in many diseases.
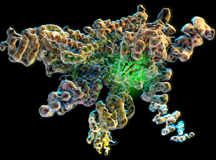
The new research is the first study in almost 20 years to solve a large component of the proteasome at near-atomic resolution. Lander said the breakthrough was possible with recent advances in cryo-electron microscopy (EM), an imaging technique in which a sample is bombarded with an electron beam, producing hundreds of thousands of protein images that can be consolidated into a high-resolution structure.
Using cryo-EM, scientists investigated part of the proteasome that contains a deubiquitinase enzyme called Rpn11. Rpn11 performs a crucial function called deubiquitination, during which it cleaves molecular tags from proteins scheduled for recycling in the proteasome. This is a key step in proteasomal processing—without Rpn11, the protein tags would clog the proteasome and the cell would die.
From previous studies, scientists knew Rpn11 and its surrounding proteins latch onto the proteasome to form a sort of lid. “The lid complex wraps around the proteasome like a face-hugger in the movie ‘Alien,’” said Lander.
The lid complex can also exist separately from the proteasome—which poses a potential problem. If Rpn11 cleaves tags from proteins that haven’t gotten to the proteasome yet, those proteins could skip the recycling stage and cause disease. Scientists had wondered how nature had solved this problem.
A Guide for Future Therapies
The study provides an answer, showing the lid complex as it floats freely in cells. In this conformation, Rpn11 is carefully nestled in the crook of surrounding proteins, stabilized and inactive.
“There's a sophisticated network of interactions that pin the Rpn11 deubiquitinase against neighboring subunits to keep it inhibited in the isolated proteasome lid,” explained Corey M. Dambacher, a researcher at TSRI at the time of the study and now a senior scientist at Omniome, Inc., who was first author of the study with TSRI Research Associate Mark Herzik Jr. and Evan J. Worden of UC Berkeley.
“In order for Rpn11 to perform its job, it has to flip out of this inhibited conformation,” said Herzik.
The new study also shows that, to flip out of the conformation at the proteasome, the proteins surrounding deubiquitinase pivot and rotate—binding to the proteasome and releasing the deubiquitinase active site from its nook.
Lander called the system “finely tuned,” but said there may be ways to manipulate it. The study collaborators at UC Berkeley made small mutations to the proteins holding Rpn11 in position, and found that any small change will release the deubiquitinase, even when the lid is floating freely.
Lander said the new understanding of the mechanism that activates Rpn11 could guide future therapies that remove damaged or misfolded proteins.
“Accumulation of these toxic proteins can lead to diseases such as Parkinson’s and Alzheimer’s, as well as a variety of cancers,” Lander said. “If we can harness the proteasome’s ability to remove specific proteins from the cell, this gives us incredible power over cellular function and improves our ability to target certain cells for destruction.”
Going forward, the researchers hope to use the same cryo-EM techniques to investigate other components of the proteasome—and figure out exactly how it recognizes and destroys proteins. “There’s still a lot to learn,” said Lander.
For more information on the study, “Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition,” see http://elifesciences.org/content/early/2016/01/08/eLife.13027
This research was supported by the Damon Runyon Cancer Research Foundation (grant DFS-#07-13), the Pew Scholars program, the National Institutes of Health (grants DP2 EB020402 and R01-GM094497), the Searle Scholars Program, the National Science Foundation CAREER Program (grant NSF-MCB-1150288), the Howard Hughes Medical Institute and a National Science Foundation Graduate Research Fellowship.
Send comments to: press[at]scripps.edu