Structural Discoveries Could Aid in Better Drug Design
By Eric Sauter
F. Scott Fitzgerald once said that the test of a first-rate intelligence is the ability to hold two opposed ideas in mind at the same time and still retain the ability to function. Now, scientists from the Florida campus of The Scripps Research Institute (TSRI) have found the biological equivalent of that idea or something very close.
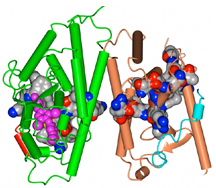
For the first time, they have uncovered the structural details of how some proteins interact to turn two different signals into a single integrated output. These new findings could aid future drug design by giving scientists an edge in fine tuning the signal between these partnered proteins—and the drug’s course of action.
“Thyroid, vitamin D and retinoid receptors all rely on integrated signals—their own signal plus a partner receptor,” said TSRI Associate Professor Kendall Nettles, who led the study with TSRI colleague Associate Professor Douglas Kojetin. “These new findings will have important implications for drug design by clearly defining exactly how these signals become integrated, so we will be able to predict how changes in a drug’s design could affect signaling.”
The study was published recently in the journal Nature Communications.
Using a number of complementary technologies, including nuclear magnetic resonance (NMR), X-ray crystallography and hydrogen/deuterium exchange (HDX) mass spectrometry from the laboratory of Scripps Florida colleague Chair of the Department of Molecular Therapeutics Patrick R. Griffin, the scientists were able to determine the mechanism through which two signaling pathways become integrated.
The study focused on a small subset of nuclear receptors, a large family of proteins that regulate gene expression in response to signals from various binding partners, including steroids and fats. Once receptors sense the presence of these binding partners, they send out new signals that initiate other cellular processes.
“Nuclear receptors bind different types of molecules, and some of these receptors physically interact with each other to integrate different signals,” Kojetin said. “Earlier studies basically accepted this without any structural evidence for communication between receptors. This is the first time that anyone has looked at what’s actually going on at the atomic level.”
In addition to Nettles, Kojetin and Griffin, authors of the study, “Structural Mechanism for Signal Transduction in RXR Nuclear Receptor Heterodimers,” include Edna Matta-Camacho, Travis S. Hughes, Sathish Srinivasan, Jerome C. Nwachukwu, Valerie Cavett, Jason Nowak, Michael J. Chalmers, David P. Marciano and Theodore M. Kamenecka of TSRI; Andrew I. Shulman of the University of California, Irvine; Mark Rance of the University of Cincinnati; and John B. Bruning of The University of Adelaide. See http://www.nature.com/ncomms/2015/150820/ncomms9013/full/ncomms9013.html
The work was supported by the National Institutes of Health (grants DK101871, GM114420, GM063855, RR019077, RR027755, MH084512, GM084041, RR027270 and CA132022), the Frenchman’s Creek Women for Cancer Research, the James and Esther King Biomedical Research Program, the Florida Department of Health and the State of Florida.
Send comments to: press[at]scripps.edu