Research Interest
Double-strand break (DSB) Repair Pathways and Cancer Connection
Major DSB Repair Pathways
DSBs can be repaired by different pathways. Non-homologous end joining (NHEJ) and homologous recombination (HR) are two major pathways to repair DSBs. NHEJ is Ku-dependent and ligates DSB ends without using a template. HR uses homologous sequences as repair templates and is considered as the most conserved repair mechanism.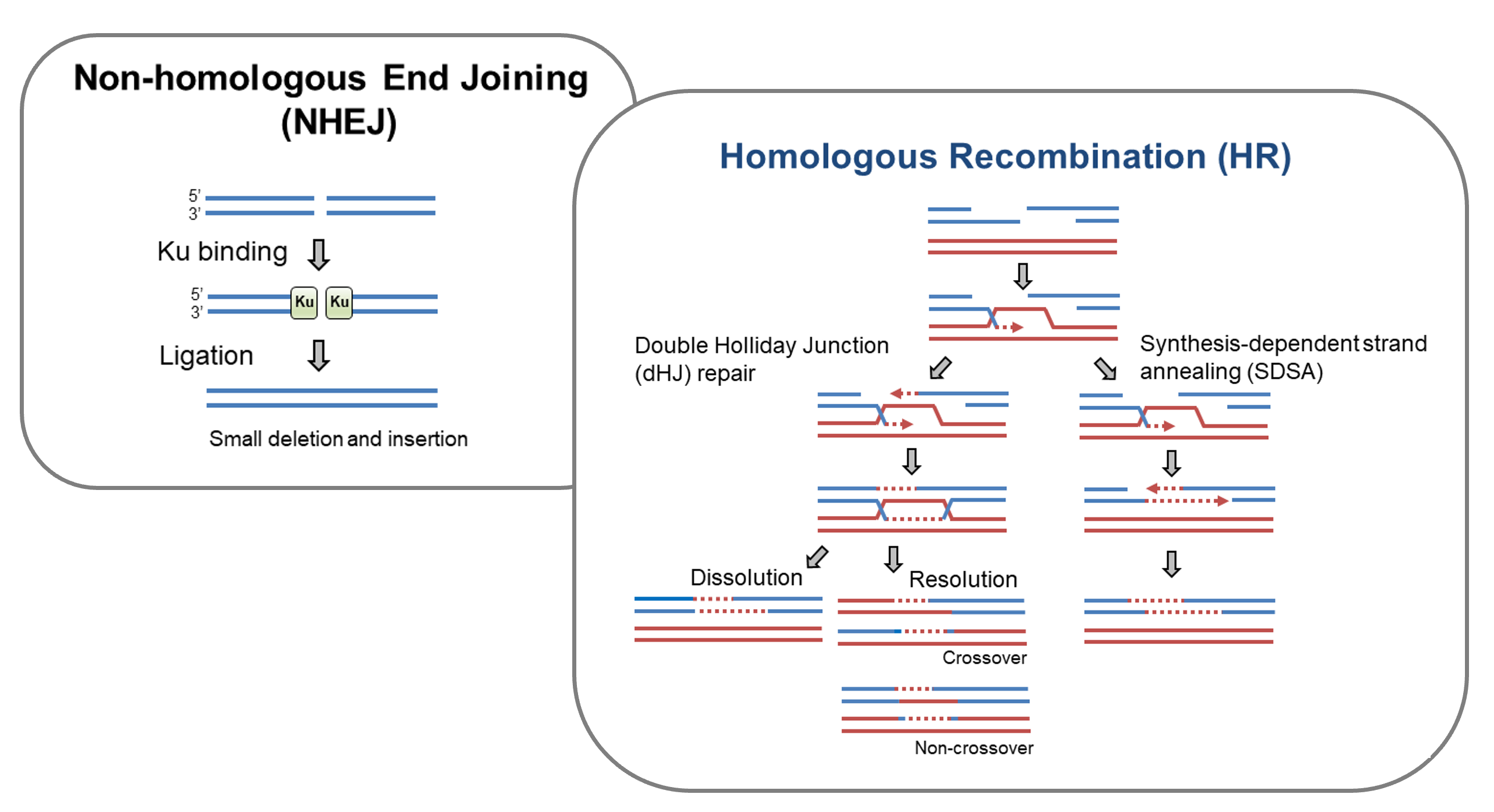
Alternative DSB Repair Pathways
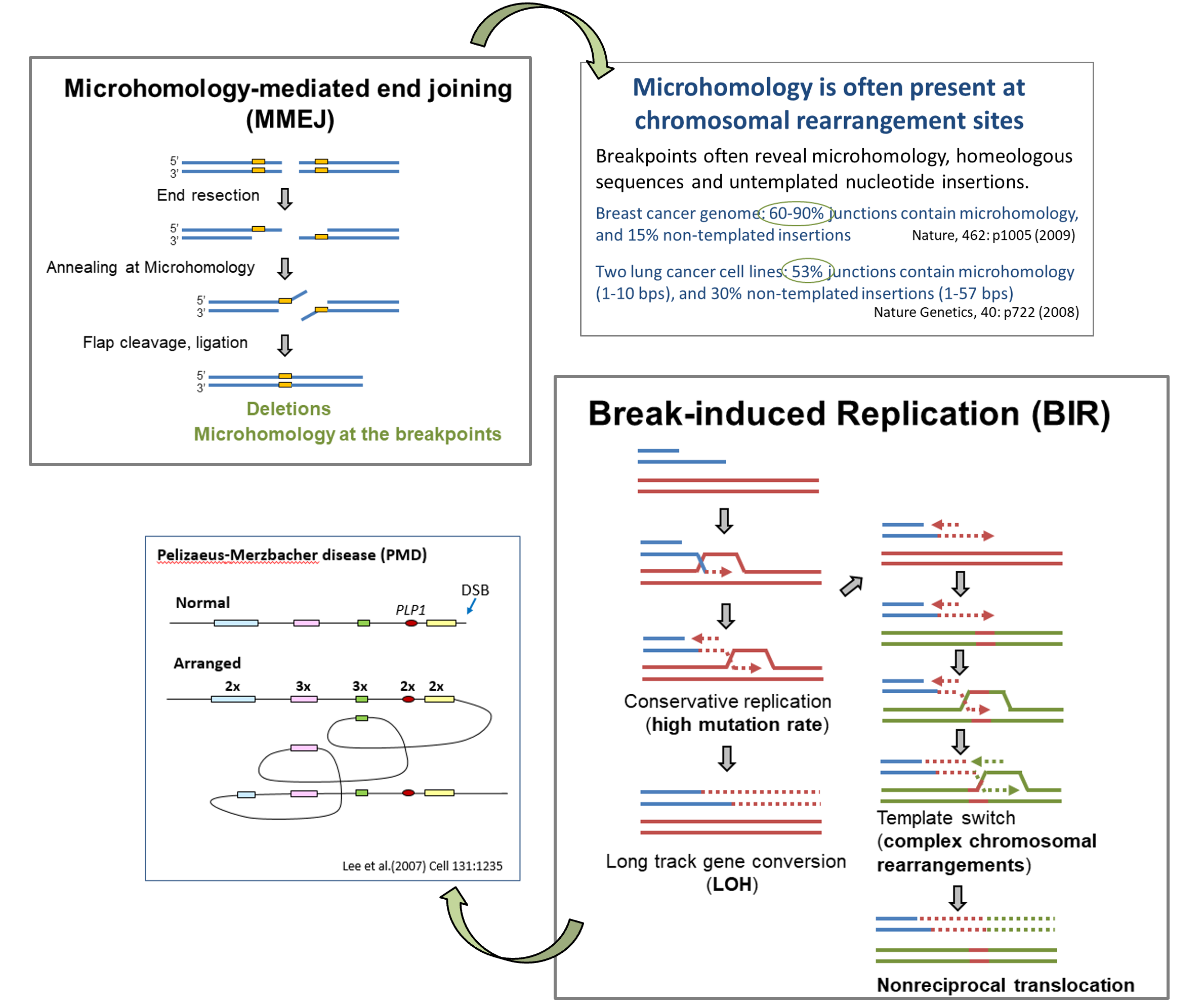
Besides HR and NHEJ, there are other alternative DSB repair pathways which are often error-prone. For instance, microhomology-mediated end joining (MMEJ) ligates DSB ends at microhomology sites and inevitably causes deletions at the repair junctions. Break-induced replication (BIR) is a variation of HR and is used when homology is present only at one side of the break. BIR exhibits conservative replication, long gene conversion track
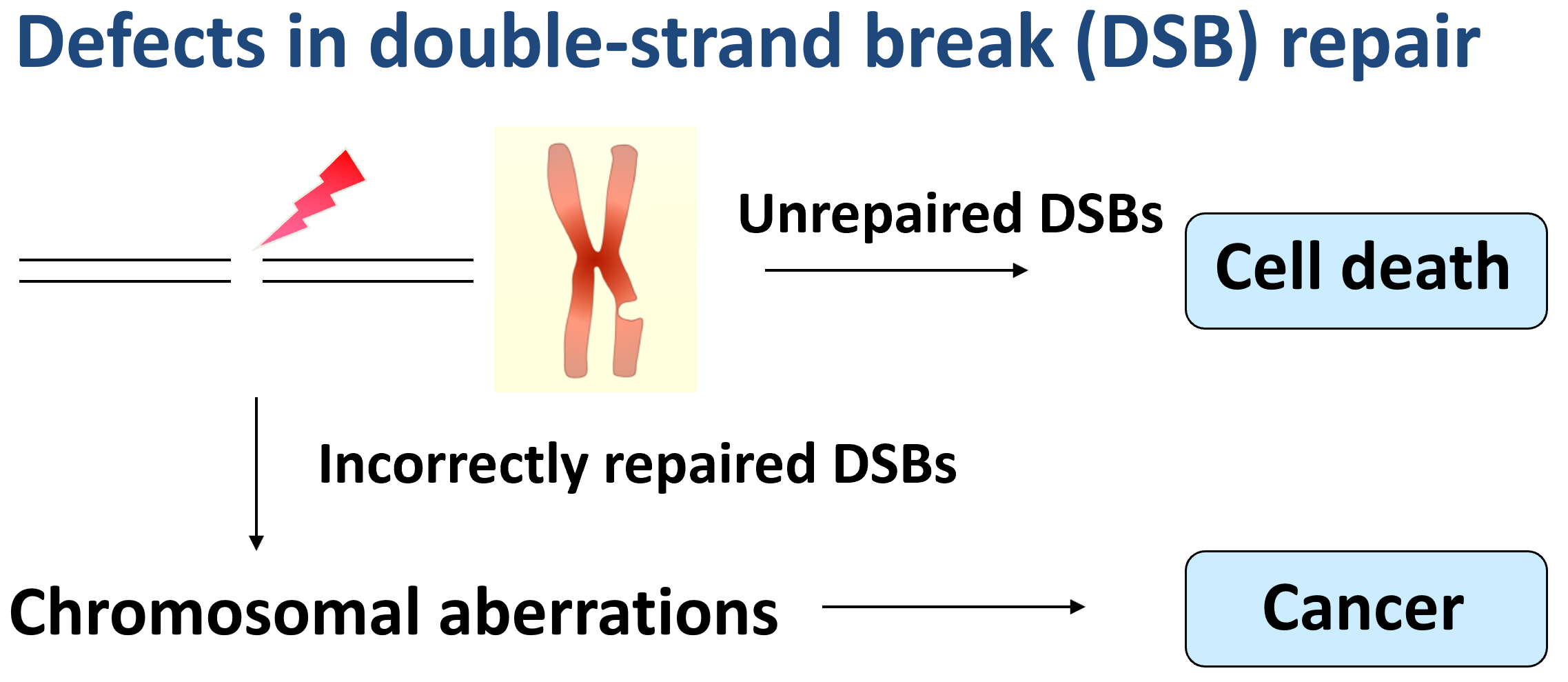
 Cancer connection
Cancer connection
Genome instability caused by DSB repair deficiency is a driving force for tumorigenesis. Cancer cells likely activate or boost alternative DSB repair pathways affording growth advantage to cope with oncogenic replication stress. Whole genome sequencing has revealed common features of gross chromosomal rearrangements associated with cancers. For instance, microhomology is often present at chromosomal breakpoints in cancer cells, and thus MMEJ is likely involved in cancer-associated chromosomal rearrangements. Complex chromosomal rearrangement and copy number variation also prevail in the cancer genome, involving replicative mechanisms to copy DNA sequences from other places of the genome. This is reminiscent of the features of BIR which could contribute to increased mutation rate, loss of heterozygosity (LOH) and copy number variation that are often found in genomic disorders and cancer cells.
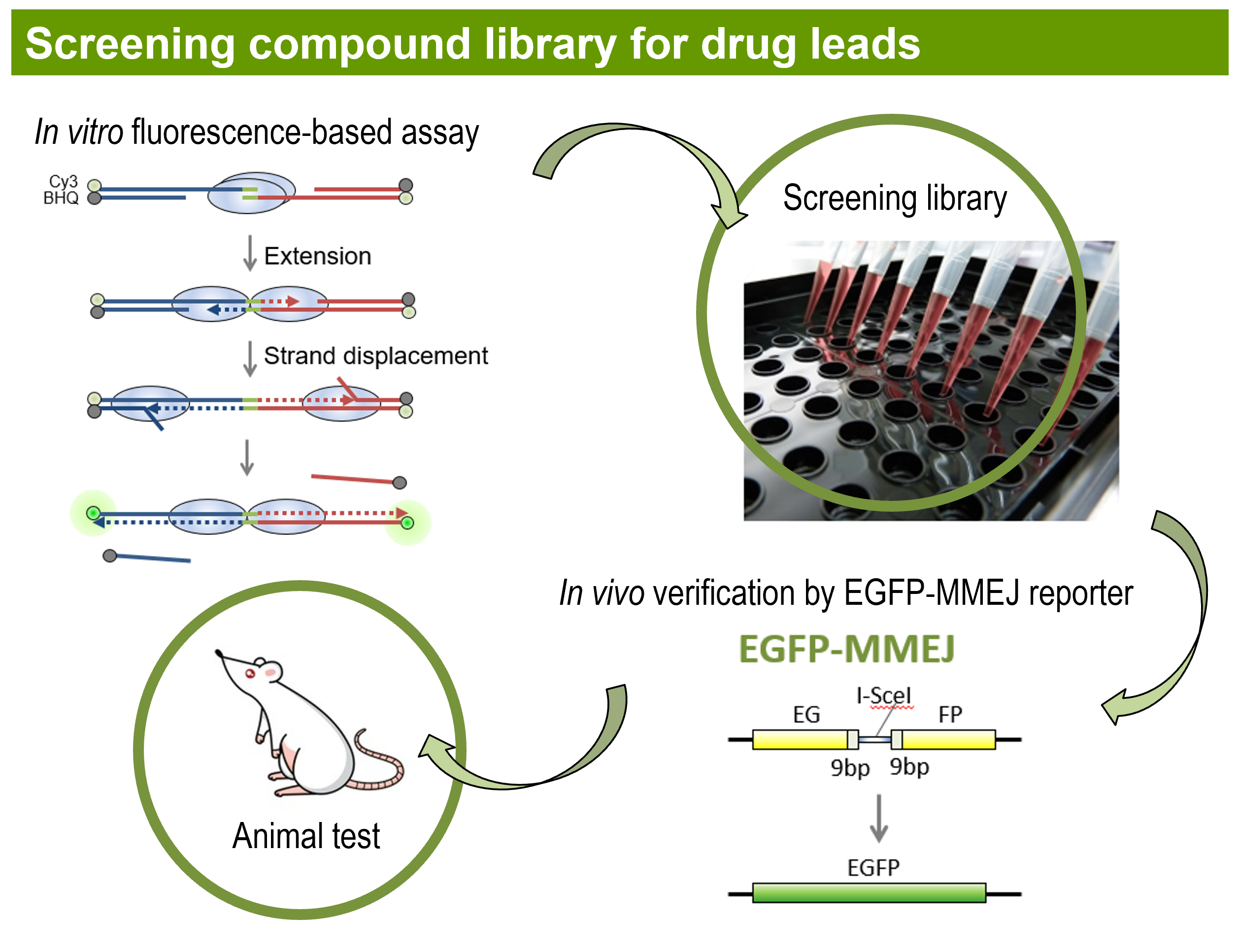
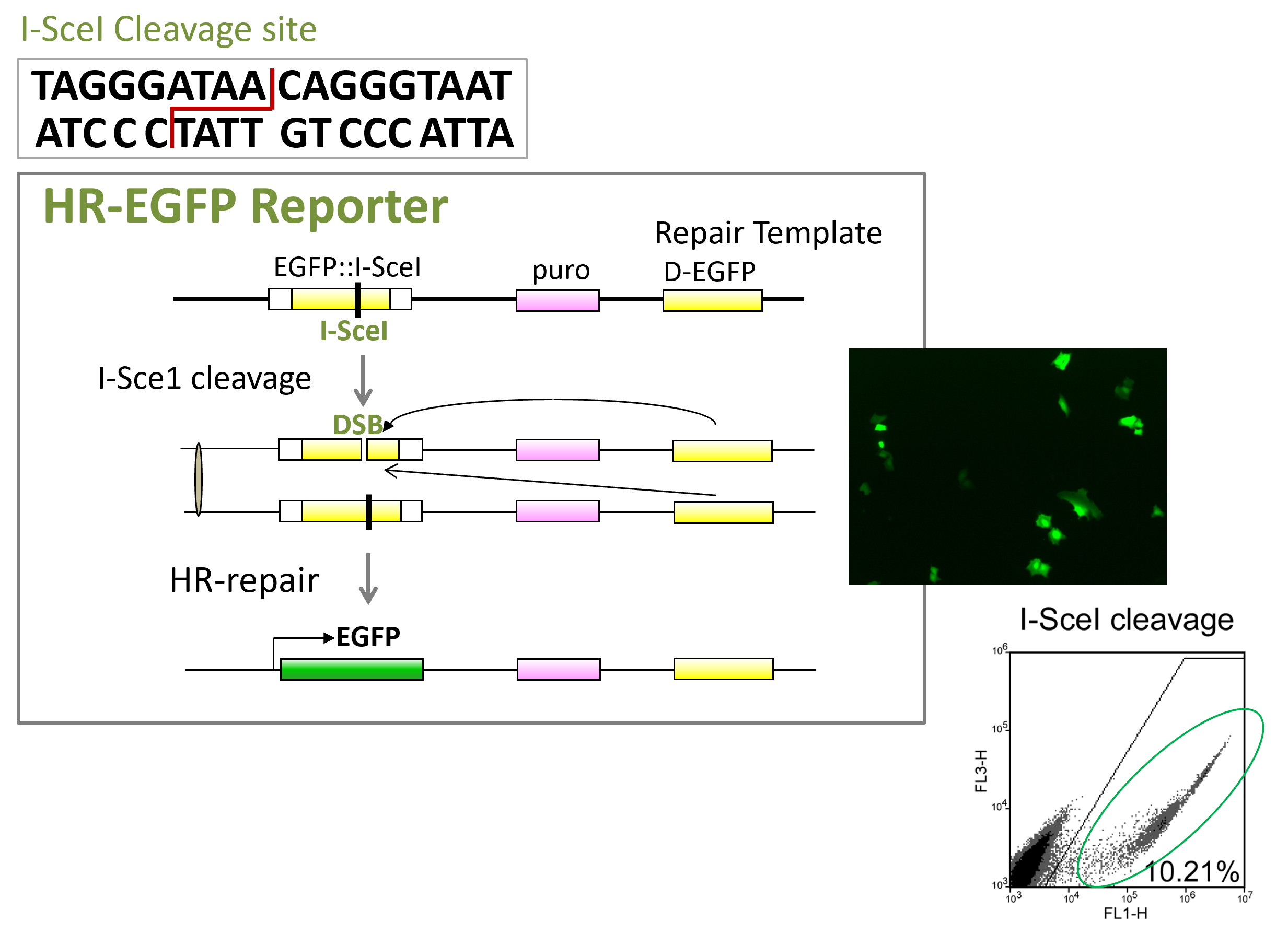
EGFP-Based DSB Repair Reporters
The challenge of studying DSB repair mechanisms in mammalian cells is the lack of sensitive and reliable DSB repair assays. We developed novel EGFP-based repair reporters allowing systematic characterization and mechanistic study of different DSB repair pathways. We use a mega-endonuclease I-SceI to create a single DSB or adapt CRISPR system to generate DSBs or nicks in the reporters, and determine the repair efficiency by FACS analysis of green cells. Such EGFP-based assay systems can be developed for CRISPR library screening and small molecule screening.
 Study Genetic and Epigenetic regulation of DSB repair.
Study Genetic and Epigenetic regulation of DSB repair.
Mutations in damage checkpoint genes and DSB repair genes are often associated with inheritable human diseases which are predisposed to cancer. BRCA1 is a breast cancer suppressor and a BRCA1 mutation carrier has 60-80% chance of a life time risk of developing breast cancer. Nbs1-, Mre11-, CtIP- and Fanconi anemia-related human diseases also exhibit genome instability and are prone to cancer. We study the role of these tumor suppressor genes in DSB repair,
Epigenetic modification and chromatin remodeling play important roles in DSB repair. We study posttranslational modifications such as phosphorylation, ubiquitination and sumoylation in the regulation of DSB repair. We also study chromatin-remodeling such as SWI/SNF complexes in DNA repair in a chromatin environment.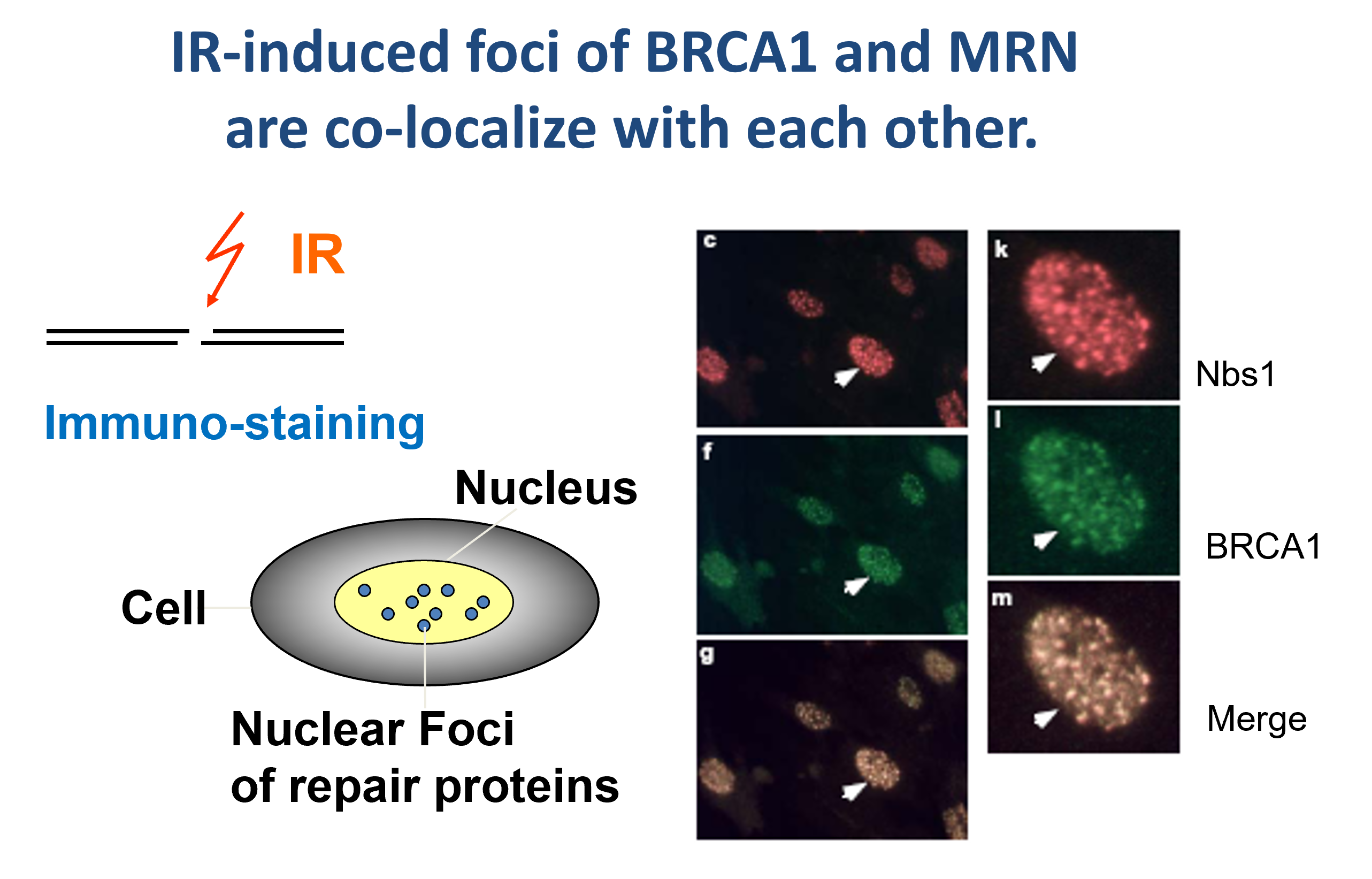
Study the mechanisms to maintain genome stability at chromosomal loci with secondary structures.
DNA sequences prone to secondary structure formation are often hot spots for chromosomal rearrangements, and many of these structures are associated with neurological diseases and cancer. Common fragile sites (CFSs) are specific chromosomal regions that are more easily to break down than other places in the genome, especially under replication stress. We identified several clusters of extremely AT-rich DNA sequences at CFSs, which are genetically unstable causing DSB formation. G-quadruplex (G4) is abundantly present in the human genome, which 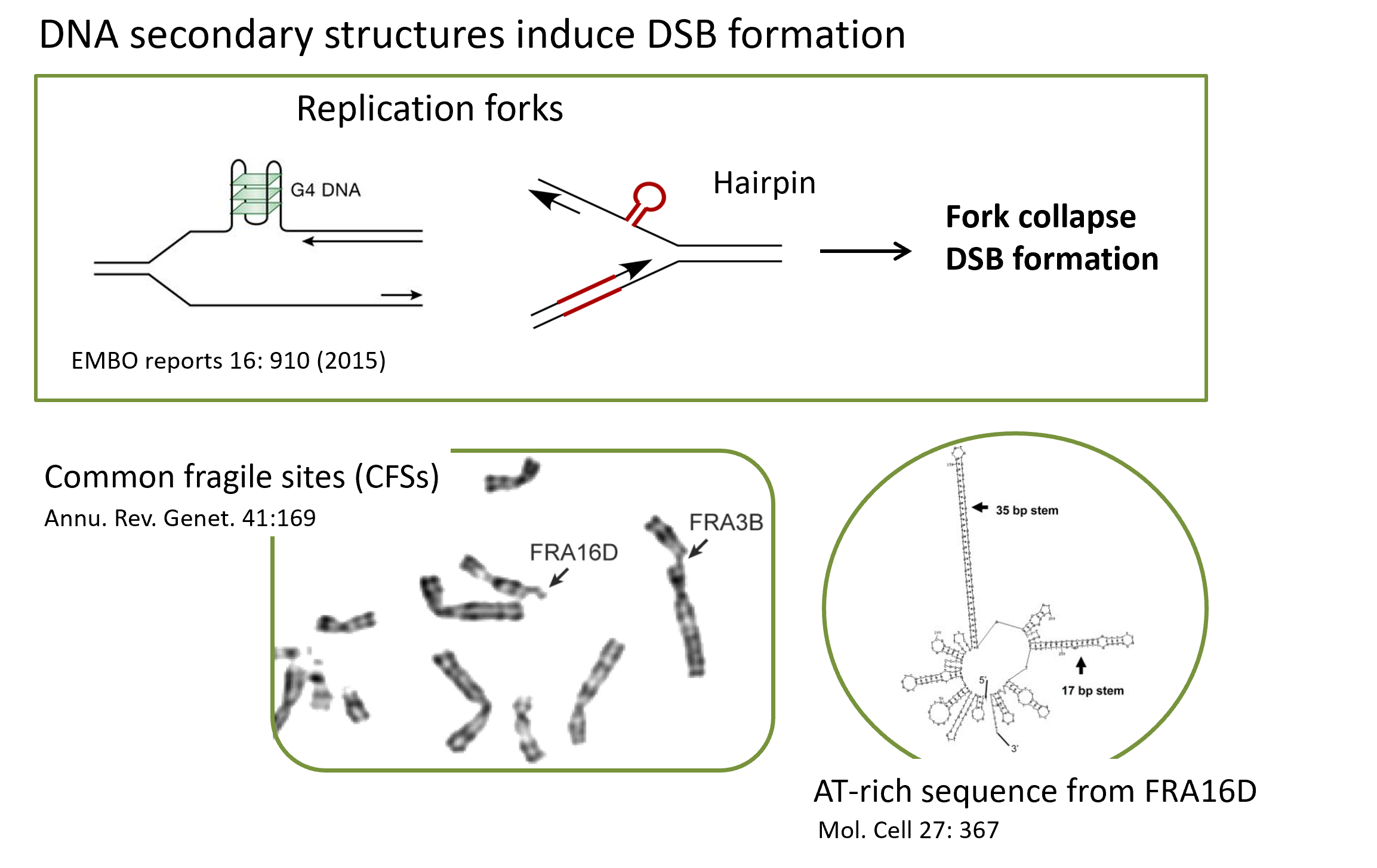
Targeting DSB repair pathways for cancer treatment.
Synthetic lethality: When a combination of deficiencies in the expression of two or more genes leads to cell death, whereas a deficiency in only one of these genes does not.
Although tumor cells proliferate fast, they often have defects. For instance, they are often defective in one or more damage checkpoint and DNA repair pathways. Through
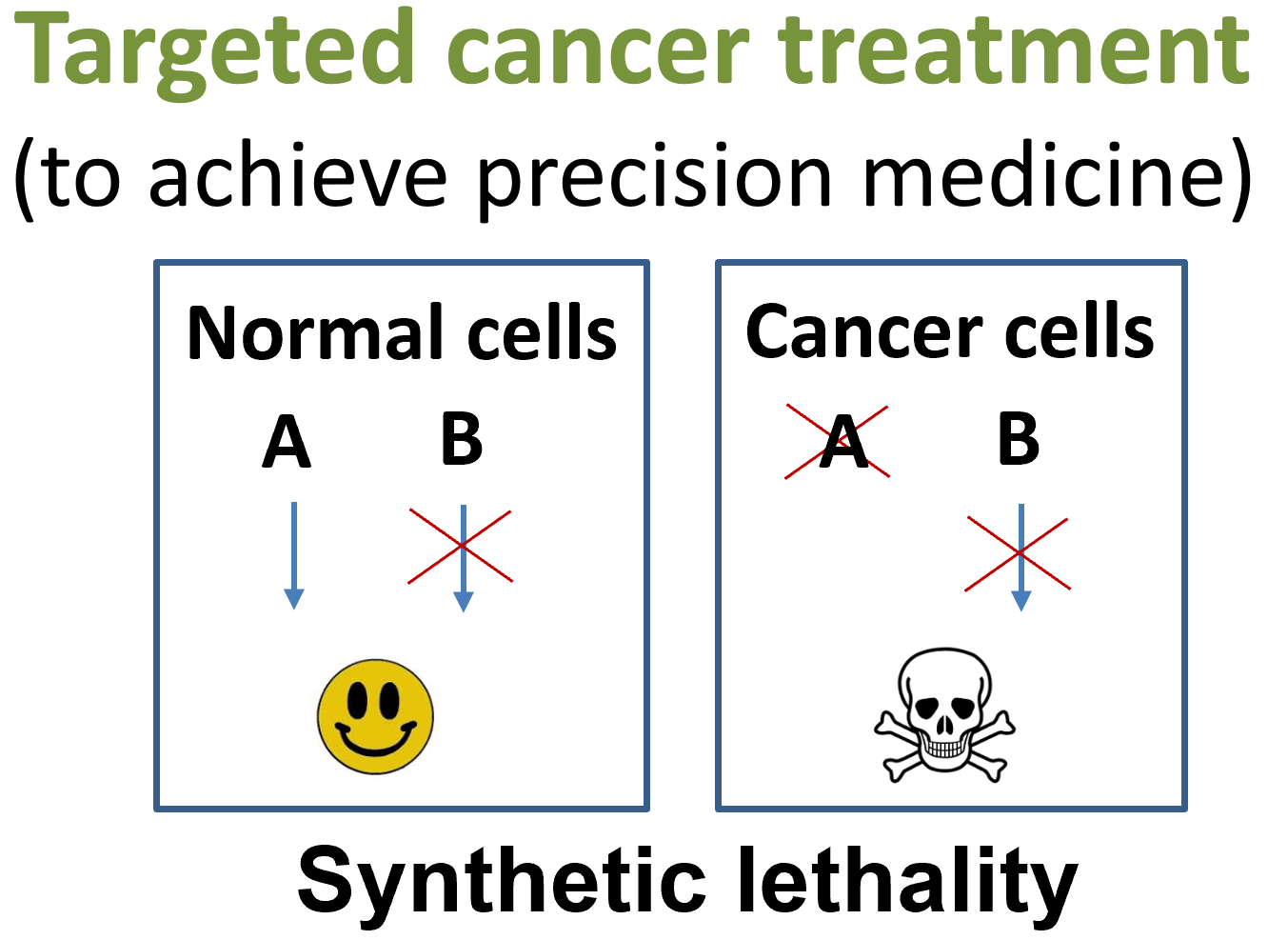 CRISPR library screening: We aim to identify synthetic lethality pairs of human genes in checkpoint and DSB repair pathways.
CRISPR library screening: We aim to identify synthetic lethality pairs of human genes in checkpoint and DSB repair pathways.
Small molecule library screening: We screen for small molecule inhibitors towards potential targets that can be used for targeted cancer treatment in