The researchers were able to monitor the progress of the transcription of ribosomal RNA from DNA, and observed that it was followed quickly by the binding of a ribosomal protein, S15, to the newborn RNA transcript. They recorded this process simultaneously for hundreds of distinct ribosome-assembly events. Credit: Williamson lab
New technique allows study of large molecule assembly in real time
December 18, 2018
LA JOLLA, CA – A team co-led by scientists from Scripps Research has demonstrated a new microscopy technique for studying complex biological processes at the molecular level. The researchers used the technique to record in real time the copying out, or transcription, of DNA into RNA, and the subsequent binding of a protein to the RNA transcript to begin the assembly of a large molecule called a ribosome.
The technique, described in a paper on November 30 in Nature Communications, paves the way for the study of a variety of RNA- and protein-related molecular interactions in unprecedented detail and under conditions like those in living cells.
“We can apply this approach to many biological problems,” says study lead author Olivier Duss, PhD, a postdoctoral fellow in the Department of Integrative Structural and Computational Biology at Scripps Research.
The senior investigators for the study were James Williamson, PhD, professor at Scripps Research, and Joseph Puglisi, PhD, professor at Stanford University.
Nanoscale biological processes such as DNA transcription cannot be observed directly via ordinary microscopes, because the molecules involved are many times smaller than visible light wavelengths. Scientists therefore have had to devise indirect means of “seeing” such processes. For example, single-molecule imaging techniques use fluorescent tags on individual molecules to monitor their motions and interactions with other molecules.
One common shortcoming of single-molecule techniques is the requirement that the tagged molecules be at very low concentration—so their collective fluorescence won’t create too much background “noise” that obscures the signal from individual molecules.
“The problem is that when you want to study some biological processes, such as ribosome assembly, you have to reproduce the much higher concentration of molecules that occurs in the cell; otherwise the process won’t happen the way it does normally,” Duss says.
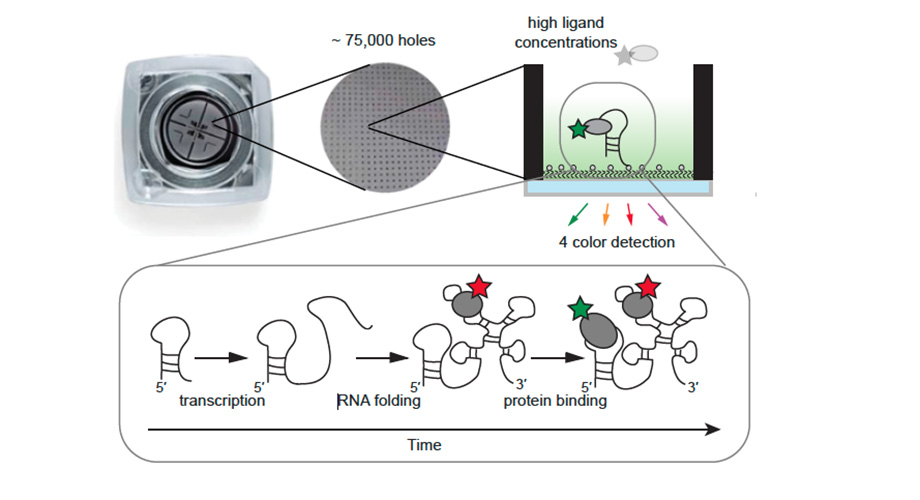
The new technique is based on a technology called zero-mode waveguide fluorescence microscopy, which allows higher, biologically realistic concentrations of the molecules that are imaged. It accomplishes this using a metal film perforated with thousands of tiny holes, or waveguides, between the illumination source and the solution of molecules. These waveguides effectively isolate individual molecules in the illumination/viewing field, even when they are at relatively high concentrations, thus greatly reducing any background noise.
In this case, the scientists attached special fluorescent tags to the molecules of interest in such a way that certain distinctive movements of the molecules would cause detectable changes in the intensity of the fluorescence. Thus, the scientists could monitor molecular interactions in real time, and do so simultaneously for the hundreds to thousands of individual sets of interacting molecules isolated by the waveguides—enabling researchers to get a statistically solid sense of what happens normally in these interactions. For each set of molecules in a waveguide the researchers could use up to four tags that fluoresce in different colors, allowing them to monitor that many different molecules in the same process at the same time.
The team demonstrated the power of the technique by using it to record an early stage of ribosome assembly. Ribosomes are molecular factories made of both RNA and protein, and perform the crucial function of synthesizing proteins in cells. Every cell needs thousands or even millions of ribosomes, and is constantly assembling new ones as old ones break down or the cell divides. At the start of ribosome assembly, the ribosomal proteins interact with the RNA while it is still being made, but how transcription and protein binding are coupled has not been well understood.
The researchers were able to monitor the progress of the transcription of ribosomal RNA from DNA, and observed that it was followed quickly by the binding of a ribosomal protein, S15, to the newborn RNA transcript. They recorded this process simultaneously for hundreds of distinct ribosome-assembly events.
The data showed that this protein-RNA binding process works well at a normal physiological temperature of 35°C, but is much less efficient at a relatively cold 20°C, apparently because the ribosomal RNA tends to misfold at that temperature as it is being synthesized during transcription.
“One reason we chose ribosomal protein S15 for our first demonstration of protein binding to a transcribing RNA is that the S15-ribosomal RNA interaction is relatively well studied, so we could check our results against what is already known,” Duss says.
Having validated their technique in this way, Duss and colleagues are now moving on to apply it to more complex and challenging biological events, including RNA folding, full ribosome assembly, and even the entire process from a ribosome’s assembly to its synthesis of new proteins.
The authors of the study, “Real-time assembly of ribonucleoprotein complexes on nascent RNA transcripts,” were Olivier Duss, Galina Stepanyuk, Annette Grot, Seán O’Leary, Joseph Puglisi, and James Williamson, of Scripps Research.
This research was made possible by grants from the National Institutes of Health (R01 GM053757, R01 GM051266), the Human Frontier Science Program (LT000628/2015-L), and the Swiss National Science Foundation (P2EZP3-152131, P300PA-160978).
For more information, contact press@scripps.edu

