Scientists Describe Cholera Protein Structure—a Target
for Vaccines and Antibiotics
By Jason Socrates
Bardi
A group of researchers from The Scripps Research Institute
(TSRI) has solved structures of a bacterial protein called
pilin, which is required for infection by pathogens that cause
human diseases like meningitis, gonorrhea, diarrheal diseases,
pneumonia, and cholera.
In the latest issue of the journal Molecular Cell,
the TSRI group reports two key structures of these pilins
and discoveries about their assembly into fibrous "pili."
Because a whole class of bacterial pathogens require the assembly
of pilin into the hair-like pilus filaments on their surface
in order for them to move around, attach to, and infect host
cells, the authors believe that this research provides essential
knowledge to help scientists develop novel antibiotics and
vaccines against these deadly and emerging bacterial diseases.
This work directly focuses on two such pathogens—Pseudomonas
aeruginosa, which causes severe lung infections in cystic
fibrosis patients, AIDS patients, and other immunocompromised
individuals, and Vibrio cholerae, which causes cholera,
a potentially fatal diarrheal disease that primarily afflicts
people in developing countries.
"Cholera," says TSRI Professor John Tainer, "is a disease
that could use better vaccines."
In the developing world and in areas with poor sewage treatment,
cholera is still a major public health problem, and can be
a deadly for children in third world countries. Although cholera
was once common in this country, modern water treatment has
virtually eliminated the disease domestically, though it is
still a concern for U.S. world travelers.
Cholera is caused by an acute intestinal infection with
the bacterium Vibrio cholerae. This usually occurs
after someone has eaten food or drank water contaminated with
the pathogen.
Cholera infections are sometimes mild, but result in watery
diarrhea, vomiting, and severe fluid loss about five percent
of the time. These cases are life-threatening and deadly where
treatment through simple rehydration with a sugar and salt
mixture is not available. There is currently no effective
vaccine available for this disease.
The Structure and How It Was Solved
Pili are key structures of the bacterium Vibrio cholerae
and several other types of bacteria. They enable the bacteria
to crawl around and stick to the intestine, lung, and other
mucosal surfaces, and to pick up foreign genes and DNA, bringing
them aboard to potentially increase the bacteria's pathogenicity.
In cholera, these pili are essential for the infection because
they allow the bacteria to clump together and form a colony
that protects them from the human immune response. This makes
pili a good target for vaccine design, since blocking them
should block the bacterium's ability to cause infection.
Tainer, who is an investigator in TSRI's Department of Molecular
Biology and The Skaggs Institute for Chemical Biology, worked
on solving the atomic structure of the pilus filaments with
Senior Research Associate Lisa Craig, and three other key
researchers—TSRI Professor Mark Yeager, computational
expert and director of graphics development at TSRI Michael
Pique, and Dartmouth Medical School Professor Ronald Taylor.
However, solving the structure of these proteins was not
easy because of their size and shape. The pili themselves
are assembled from thousands of copies of a single pilin subunit
protein stacked together to resemble a microscopic thread—they
are several hundred times longer than they are wide.
These structures are too large and flexible to be solved
with the traditional techniques of structural biology used
to study small proteins.
Yet members of the research team were aware that solving
the structures was an important goal. "If we can understand
their atomic structure, we can go after developing vaccines
that are highly specific," says Craig, who is first author
on the paper.
So in the current study, the TSRI team was creative and
combined more than one approach.
The group first solved the structure of the individual pilin
proteins from the V. cholerae bacterium using x-ray
crystallography—a technique where scientists first make
crystals of molecules like proteins or DNA and then expose
them to x-rays. The pattern of diffracted x-rays can then
be collected and analyzed to determine the structure of the
molecules in the crystal. Although a fragment of the V.
cholerae pilin protein was missing in their structure,
they were able to infer this structure by solving a full length
structure of a pilin subunit from P. aeruginosa, which
is important in infections of children with cystic fibrosis.
Craig, Yeager, and Tainer then used a technique called electron
microscopy to understand how the pilin proteins were organized
in the pilus filaments. Electron microscopy uses a beam of
electrons to magnify protein assemblies and other tiny structures
up to one hundred thousand times onto a digital camera or
a photographic plate.
The integration of x-ray crystallography and electron microscopy
allowed Craig, Pique, and Tainer to build a model of the pili
otherwise impossible at that level of molecular detail.
And the structures give new insights into how the pili assemble
and how they contribute to the pathogenesis of the bacteria—as
well as providing a unique molecular map of these proteins
that should aid in the design of new vaccines and therapeutics.

|

Professor John Tainer led a group of
researchers that solved structures in bacteria that may become
targets for drug development. Photo by Biomedical
Graphics.

Research Associate Lisa Craig is first
author on the recent paper the group published in the journal
Molecular Cell. Photo
by Jason S. Bardi.
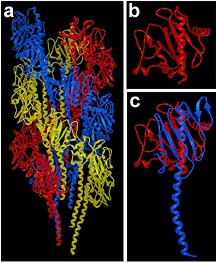
The structure and assembly of Vibrio
cholerae toxin coregulated pilus (Panel a) was based
on the crystal structures of V. cholerae pilin (red,
Panels b and c) and full length PAK pilin (blue, Panel c),
as well as crystal packing interactions and cryo-electron
microscopy of toxin coregulated pilus filaments.
Image courtesy of Lisa Craig.
|

