A Non-PCR DNA Detection Method
By Jason Socrates
Bardi
Polymerase Chain Reaction (PCR), one of the broadest technologies
to have emerged from biology in the last quarter century,
has become so commonplace across fields from archeology to
criminal investigations that it is hard to imagine what we
ever did without it.
And harder, still, to imagine why we would want to do without
it today.
Yet despite its broad application and its continued technological
improvements, PCR requires a trained technician, necessitating
either the expense of employing such a person in-house or
the time and expense of sending samples to a dedicated facility.
Now TSRI Ph.D. graduate and current research associate Alan
Saghatelian, TSRI graduate student Desiree Thayer, research
associate Kevin Guckian, and Professor Reza Ghadiri in the
Department of Chemistry have designed a non-PCR method for
detecting specific sequences of nucleic acid that may have
advantages over PCR, especially in such situations as field
work and point-of-care medicine where the technology could
be used by non-specialists. The new method is exquisitely
sensitive and quite fast, according to Ghadiri, detecting
as minute a sample as 10 femtomoles of DNA in less than three
minutes.
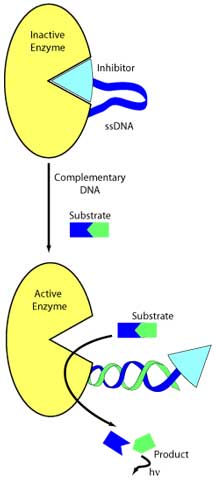
The method makes use of a detection system based on an inhibitor–DNA–enzyme
complex. Specifically, the complex is composed of an enzyme,
a single-stranded piece of DNA covalently attached to the
enzyme, and, at the end of this DNA strand, an "intramolecular"
inhibitor. The complex is able to "detect" pieces of DNA that
are complimentary to its single strand of DNA.
When complimentary DNA is not present, the single
strand of DNA in the complex is flexible enough that it can
loop around, allowing the inhibitor to occupy the binding
site of the enzyme. But when complimentary DNA is present,
the complimentary DNA forms a duplex with the complex's single
strand—straightening out the DNA—and the inhibitor
at the end on this duplex can no longer occupy the enzyme's
binding site, enabling the enzyme to cleave its substrate.
Ghadiri and his colleagues selected a fluorophoric substrate
so that this cleavage releases energy in the form of easily
detected fluorescence, signaling the presence of complimentary
DNA. The sensitivity of the method comes from the fact that
the system is self-amplifying. Any one molecule of DNA that
hybridizes to one complex turns on that one enzyme, which
can then do multiple turnovers of the substrate.
To read the article, "DNA Detection and Signal Amplification
via an Engineered Allosteric Enzyme" by Alan Saghatelian,
Kevin M. Guckian, Desiree A. Thayer, and M. Reza Ghadiri (J.
Am. Chem. Soc., 125 (2), 344 -345, 2003), please
see:
http://pubs.acs.org/cgi-bin/abstract.cgi/jacsat/2003/125/i02/abs/ja027885u.html.

|
TSRI scientists
have designed a new method to rapidly and sensitively detect
specific sequences of DNA.
|

