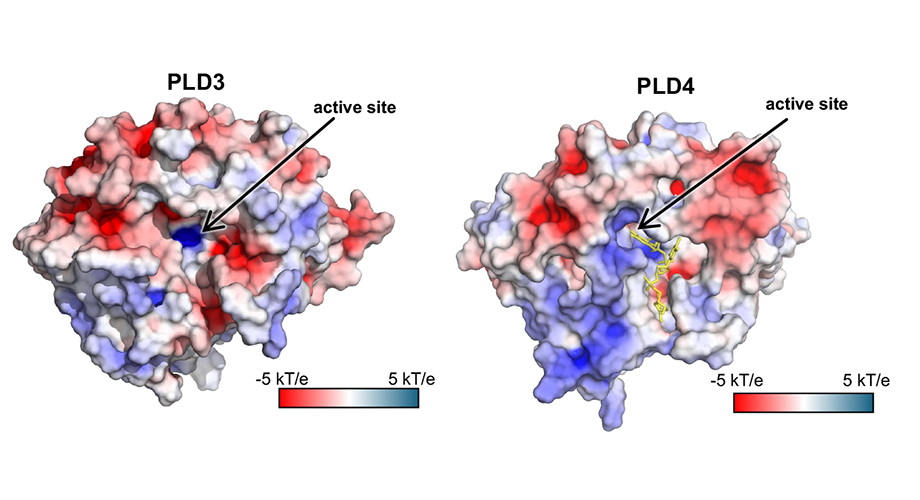
Structural models of PLD3 and PLD4, enzymes that degrade nucleic acids in the cytoplasm. The enzymes' active (or binding) sites are indicated with black arrows. Credit: Scripps Research
New enzyme models reveal key disease insights
Scripps Research scientists create near-atomic-level structural models of enzymes associated with autoimmune and inflammatory diseases, including lupus and Alzheimer’s.
April 02, 2024
LA JOLLA, CA—When nucleic acids like DNA or RNA build up in a cell’s cytoplasm, it sets off an alarm call for the immune system. Enzymes usually clear these nucleic acids before they cause an issue, but when these enzymes don’t work and the immune system gets called in, it can lead to autoimmune and inflammatory diseases.
In a new study published on March 26, 2024 in the journal Structure, Scripps Research scientists present the structures of two of these nucleic acid-degrading enzymes—PLD3 and PLD4. Understanding these enzymes’ structures and molecular details is an important step towards designing therapies for the various diseases that arise when they malfunction, which include lupus erythematosus, rheumatoid arthritis and Alzheimer’s’ disease.
“These enzymes are important for cleaning up the cellular environment, and they also set the threshold for what is considered an infection or not,” says senior author David Nemazee, PhD, professor in the Department of Immunology and Microbiology at Scripps Research. “I'm hoping someday we may be able to help patients based on this information.”
Enzymes are proteins that speed up chemical reactions of specific molecules called “substrates”. In the case of PLD3 and PLD4, the substrate is a single strand of RNA or DNA, which the enzymes break down nucleotide-by-nucleotide.
The team used X-ray crystallography to build near-atomic-scale models of the PLD3 and PLD4 in multiple states or situations, allowing them to examine how they function over the course of the catalytic reaction. This included when the enzymes were resting, or when they were actively bound to a substrate.
“These models allow us to visualize PLD3 and PLD4 very clearly and with high resolution, so we know how the atoms interact, meaning we can deduce how the enzymes work,” says first author Meng Yuan, an Institute Investigator in the Department of Integrative Structural and Computational Biology at Scripps Research.
The structural analyses revealed that PLD3 and PLD4 are structurally similar and that they degrade nucleic acids in a very similar fashion. Both enzymes degrade nucleic acids via a two-step process.
“We call this process a two-step catalysis: link-and-release,” says Yuan. “In the first step, the enzyme cleaves and links DNA strand and separates a single ‘brick’ or nucleotide from the rest of the strand, and in the second step it releases the nucleotide.”
Because the enzymatic reaction happens so quickly—within milliseconds—the researchers needed to use an alternative substrate in order to visualize the enzymes’ structure during catalysis. To do this, they incubated the enzymes together with a molecule that looks very similar to the substrate that the enzyme usually degrades, but that the enzymes degrade much more slowly.
This method uncovered a previously unknown function for one of the enzymes: in addition to cleaving nucleotides from single-stranded RNA and DNA, the enzymes also showed phosphatase activity.
“I think it's amazing that the crystal structure told us about this phosphatase activity,” says Nemazee. “To discover new enzymatic activity is unheard of in structural biology. It's only because Meng was able to solve such an amazingly accurate and detailed structure that he could inform us about this extra enzymatic activity that we had no idea about.”
After they had elucidated PLD3 and PLD4’s usual structure, the researchers examined the structure models of variants that are associated with diseases including Alzheimer’s and spinocerebellar ataxia. These analyses revealed that some of these variants had decreased enzymatic capability, while others—including a mutation associated with late-onset Alzheimer’s—appeared to be more active.
“Some of our data suggests that one of these Alzheimer's-associated enzyme variants might function better, which was a surprise to me, but it also may be less stable and more easily aggregated,” says Nemazee.
The researchers plan to continue investigating the structure and function of these enzymes. Their next steps include exploring possible ways of inhibiting the enzymes in scenarios where they are overactive, and they also plan to investigate the possibility of replacing the enzymes in people who carry non-functional (or non-working) versions.
In addition to Nemazee and Yuan, authors of the study, “Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4,” were Linghang Peng, Deli Huang, Amanda Gavin, Fangkun Luan, Jenny Tran, Ziqi Feng, Xueyong Zhu, Jeanne Matteson, and Ian Wilson of Scripps Research.
This study was supported by the National Institutes of Health (grants R01AI142945 and RF1AG070775) and Skaggs Institute for Chemical Biology at Scripps Research.
For more information, contact press@scripps.edu

