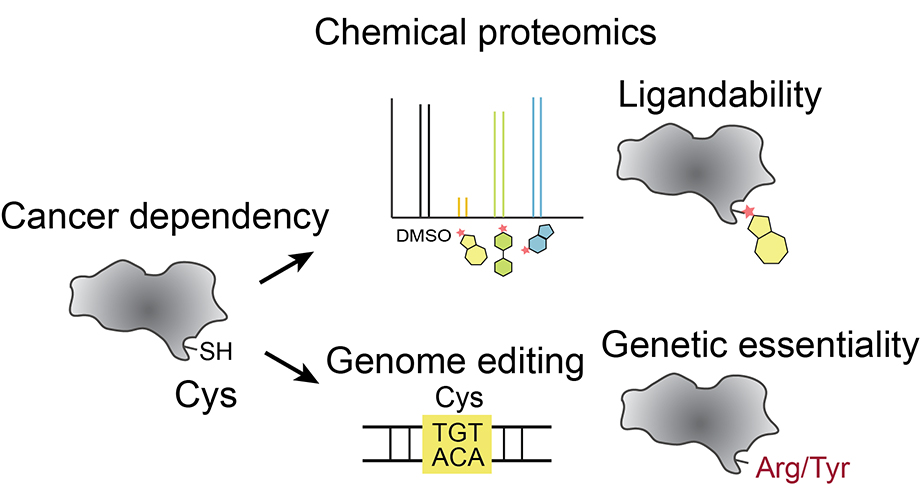
Scripps Research scientists combined two methods to study cysteine residues in proteins previously linked to cancer. Using chemical proteomics (top), they learned which cysteines were accessible to drugs. Using genome editing (bottom), they analyzed the functional consequences of altering each cysteine. Together, they suggest which cysteines may be effective cancer drug targets. Credit: Scripps Research
New cancer drug targets uncovered using base editing and chemical proteomics
Combining precise genome engineering and protein activity profiling to methodically study thousands of cancer genes, Scripps Research scientists were able to pinpoint promising cancer drug targets.
October 05, 2023
LA JOLLA, CA—Searching for new ways to block the growth of cancer cells is like looking for a needle in a haystack. Tumor cells rely on thousands of proteins to function, but only a few of those proteins can be precisely targeted by drugs to treat cancer safely and effectively. Now, a team at Scripps Research and the Broad Institute of Harvard and MIT has spearheaded a new method to hone in on new drug targets most likely to impact multiple cancers.
The research, published in Nature Chemical Biology on October 2, 2023, used a precise gene editing approach to alter more than 13,000 possible drug targets and see which edits affected cell growth. Integrating these data with chemical proteomic information pointed toward hundreds of possible drug targets, many of them never pursued in the past.
“This approach integrates two innovative methods that, together, form a target and drug discovery strategy for nominating new cancer treatments,” says senior author Benjamin Cravatt, PhD, the Norton B. Gilula Chair of Chemical Biology and chemistry professor at Scripps Research. “It provides preclinical information about which of the thousands of different protein sites are most likely to impact cancer cell growth.”
Over the past decade, research chemists and pharmaceutical companies have become excited about a class of drug that works by permanently binding to cysteines—one of the twenty amino acids that, in various combinations, make up all human proteins. The unique reactive chemistry of cysteines makes them easy and ideal drug targets, but because there are hundreds of thousands of cysteines scattered among human proteins, narrowing down which ones to target with drugs is extremely difficult. Even among the few thousand proteins that have been identified as critical to cancer cell growth, there are still more than 13,000 cysteines.
“We needed a way to narrow down the list to cysteines that have an important functional impact on cancer-relevant proteins,” says Haoxin Li, a postdoctoral research associate at Scripps Research and first author of the new paper.
As a graduate student, Li worked at the Broad Institute under the supervision of Stuart Schreiber, who uses human genetic variation to inform drug discovery. Inspired by this approach, Li wondered how he could combine precise genome engineering techniques and cutting-edge chemical proteomic tools to find the next generation of cancer therapeutics. When Li joined Cravatt’s lab at Scripps Research, he launched a collaboration with Broad Institute core member David Liu, whose lab spearheaded the development of base editing (a method to precisely change DNA letters). Li then used base editing to create targeted amino acid changes in cancer cells. By introducing a variety of cysteine-targeted mutations, he hypothesized he could learn more about which cysteines were most relevant to cancer cells.
In the new paper, Li, Cravatt and colleagues edited 13,800 spots on more than 1,750 genes previously linked to cancer cell survival. In each case, the edit targeted a cysteine on the corresponding protein. They then tested how well cancer cells with the mutation grew. Moreover, they integrated their findings with new data on the “druggability” of these cysteines. They ultimately found that about 160 of the druggable cysteines, when edited, impacted cancer cell growth—suggesting that drugs binding to these cysteines could potentially work to treat cancer.
Among the edits with the largest impacts was a change to the cancer-dependency protein TOE1. Although TOE1 is known to play an important role in trimming the ends of the cell’s RNA molecules, it had not been studied as a cancer drug target. However, Li and Cravatt’s team showed that small molecules could be harnessed to target this “Achilles' heel” of cancer cells.
“We have found compounds targeting TOE1 and showed they can block its normal activity through a fascinating mechanism that is likely to impact cancer cell growth,” says Li.
More research is needed to show whether a drug targeting TOE1 could be useful in human patients, but the initial results suggest that the approach of editing cysteines was effective in predicting which cysteines could work as drug targets.
The researchers now plan to follow-up on other novel targets that their experiments revealed. They also say that the next generation of chemical genetic approaches are currently under development to study druggable cysteines in diseases other than cancer.
In addition to Cravatt and Li, authors of the study, “Assigning functionality to cysteine by base editing of cancer dependency genes,” include Jarrett Remsberg, Sang Joon Won, Kristen DeMeester, Evert Njomen, Daisuke Ogasawara and Bruno Melillo of Scripps; Kevin Zhao, Tony Huang, Stuart Schreiber and David Liu of the Broad Institute of Harvard and MIT; Bingwen Lu and Gabriel Simon of Vividion Therapeutics; and Tiantai Ma and Jens Lykke-Andersen of UC San Diego.
This work was supported by funding from the National Institutes of Health (R35 CA231991, U01 AI142756, RM1 HG009490, R35 GM118062, R35 GM118069), the Damon Runyon Cancer Research Foundation (2406-20), the Jane Coffin Childs Fund, the Mark Foundation for Cancer Research and the Howard Hughes Medical Institute.
For more information, contact press@scripps.edu

