
Postdoctoral associate Manish Sharma and Associate Professor Srinivasa Subramaniam consult in their lab at Scripps Research in Jupiter, Florida.
Protein that polices mitochondria in brain’s striatum may underlie Huntington’s selective damage, study finds
November 07, 2019
JUPITER, FL — Parkinson’s, ALS and Huntington’s disease all share a curious feature: The genetic mutations underlying the diseases appear in all cells, yet only specific brain regions, or cell types, initially die from those mutations.
A new study from Scripps Research published online in the journal the Proceedings of the National Academy of Science (PNAS) offers a possible reason for such tissue-specific vulnerability in the case of Huntington’s disease. The discovery, from the lab of Scripps Research Neuroscientist Srinivasa Subramaniam, PhD, may offer clues to similarly isolated tissue vulnerability in other degenerative brain diseases, and may point to new therapeutic approaches.
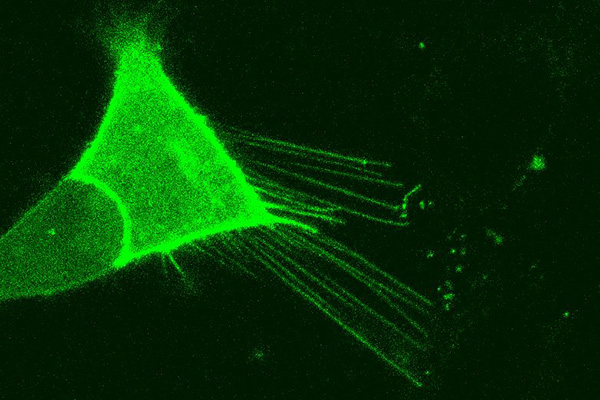
The Subramaniam lab's previous work showed that the Rhes protein is responsible for extending nanotube filaments to neighboring neurons.
The region of the brain associated with voluntary movement, called the striatum, is under attack in Huntington’s disease. Huntington’s affects an estimated 1 in 10,000 people in the United States. Its symptoms, including slowness, muscle jerks, loss of coordination, slurred speech and difficulty eating and swallowing, usually appear between ages 30 and 50. There is no treatment.
The disease, usually inherited, is caused by a toxic number of repeats of three nucleotides in one gene, called HTT, which encodes a protein called Huntingtin. But why does the Huntingtin mutation preferentially lead to the death of striatal cells but not other brain cells? Neuroscientist Srinivasa Subramaniam, PhD, and his team looked at a normal process called mitophagy, which involves removal of damaged or unnecessary mitochondria on a regular basis. Mitochondria are organelles that convert sugar into usable chemical energy, and their number per cell fluctuates constantly. The process of their removal is not well-understood in all cell types.
Subramaniam and his team discovered that in the striatum, a protein called Rhes forms a complex with another protein, Nix, to help maintain the optimal number of mitochondria. In healthy striatal cells, they found that Rhes conducts surveillance of the neurons’ mitochondria. In a model of Huntington’s, if Rhes detects damaged mitochondria, it moves quickly to recruit factors that engulf and dissolve the organelle. If too many mitochondria are wiped out, however, this leads to neuronal death, and thus a protective cellular mechanism transforms into predator.
“In normal conditions, all those parts are recycled. But in Huntington’s so many mitochondria are lost that the cell just dies,” Subramaniam says.
Rhes is disproportionately present in the striatum, so “this gives you a possible mechanism for selective vulnerability.” Subramaniam adds. “So the next question is, can we target this to prevent the excess removal?”
First author Manish Sharma, PhD, a postdoctoral associate in Subramaniam’s lab, said they will look first at how Rhes recognizes damaged mitochondria.
“One of the interesting questions is how Rhes get the message or signal about bad mitochondria in the cell?” Sharma said. “This may lead us to design the inhibitors to regulate the elimination of excessive mitochondria and cell damage in brain.”
Rhes appears capable of spreading the pathology from cell to cell as well, he adds. Subramaniam’s group previously showed that Rhes builds nanotube filaments between neurons. These tubes allow the transportation of cellular cargo between cells. It appears that in Huntington’s, these tubes accelerate the spread of the disease from one cell to another, Subramaniam says.
“We always thought of these cells as independent units. But here we have found these beautiful protrusions causing interactions among cells. This really changes our thinking of how brain cells interact.”
While Huntington’s affects the striatum, in Parkinson’s disease, another brain region, the substantia nigra, degenerates first. A next step for his group will be searching for players similar to Rhes that are uniquely overexpressed in that brain region.
“It is clear there are tissue specific proteins in other areas of the brain that may be promoting the vulnerability of those cells,” Subramaniam says.
The study, “Rhes, a striatal-enriched protein, promotes mitophagy via Nix,” was published online at www.pnas.com on Nov. 1, 2019. Besides lead author Subramaniam and first author Sharma, the co-authors include Uri Nimrod Ramirez Jarquin, Oscar Rivera, Melissa Karantzis, Mehdi Eshragi, Neelam Shahani and Vishakha Sharma of Scripps Research, Florida, and Ricardo Tapia of the Universidad Nacional Autonoma de Mexico.
This research was partially supported by a training grant in Alzheimer’s Drug Discovery from the Lottie French Lewis Fund of the Community Foundation for Palm Beach and Martin Counties. This research was supported by funding from NIH/National Institute of Neurological Disorders and Stroke grant R01-NS087019-01A1, NIH/National Institute of Neurological Disorders and Stroke grant R01-NS094577-01A1, and grants from Cure Huntington Disease Initiative (CHDI) Foundation. Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (Project IN206719), also supported the work, in part.
For more information, contact press@scripps.edu

