Scientists Uncover Why Hepatitis C Virus Vaccine Has Been Difficult to Make
Researchers have been trying for decades to develop a vaccine against the globally endemic hepatitis C virus (HCV). Now scientists at The Scripps Research Institute (TSRI) have discovered one reason why success has so far been elusive.
LA JOLLA, CA—October 24, 2016—Researchers have been trying for decades to develop a vaccine against the globally endemic hepatitis C virus (HCV). Now scientists at The Scripps Research Institute (TSRI) have discovered one reason why success has so far been elusive.
Using a sophisticated array of techniques for mapping tiny molecular structures, the TSRI scientists analyzed a lab-made version of a key viral protein, which has been employed in some candidate HCV vaccines to induce the body’s antibody response to the virus. The researchers found that the part of this protein meant as the prime target of the vaccine is surprisingly flexible. Presenting a wide variety of shapes to the immune system, it thus likely elicits a wide variety of antibodies, most of which cannot block viral infection.
“Because of that flexibility, using this particular protein in HCV vaccines may not be the best way to go,” said co-senior author TSRI Associate Professor Mansun Law.
“We may want to engineer a version that is less flexible to get a better neutralizing response to the key target site and not so many off-target responses,” said co-senior author Ian A. Wilson, TSRI’s Hansen Professor of Structural Biology and a member of the Skaggs Institute for Chemical Biology at TSRI.
The report, published online ahead of print by the Proceedings of the National Academy of Sciences the week of October 24, 2016, is likely to lead to new and better HCV vaccine designs.
A Great Need
A working vaccine against this liver-infecting virus is needed desperately. HCV infection continues to be a global pandemic, affecting an estimated 130 to 150 million people worldwide and causing about 700,000 deaths annually from liver diseases including cancer. Although powerful antiviral drugs have been developed recently against HCV, their extremely high costs are far beyond the reach of the vast majority of people living with HCV infection. Moreover, antiviral treatment usually comes too late to prevent liver damage; HCV infection is notorious for its ability to smolder silently within, producing no obvious symptoms until decades have passed.
The Law and Wilson laboratories have been working together in recent years to study HCV’s structure for clues to successful vaccine design. In 2013, for example, the team successfully mapped the atomic structure of the viral envelope protein E2, including the site where it binds to surface receptors on liver cells.
Because this receptor-binding site on E2 is crucial to HCV’s ability to infect its hosts, it has an amino-acid sequence that is relatively invariant from strain to strain. The receptor-binding site is also relatively accessible to antibodies, and indeed many of the antibodies that have been found to neutralize a broad set of HCV strains do so by targeting this site.
For all these reasons, HCV’s receptor-binding site has been considered an excellent target for a vaccine. But although candidate HCV vaccines mimicking the E2 protein have elicited high levels of antibodies against the receptor-binding site, these antibody responses—in both animal models and human clinical trials—have not been very effective at preventing HCV infection of liver cells in laboratory assays.
Enormous Flexibility
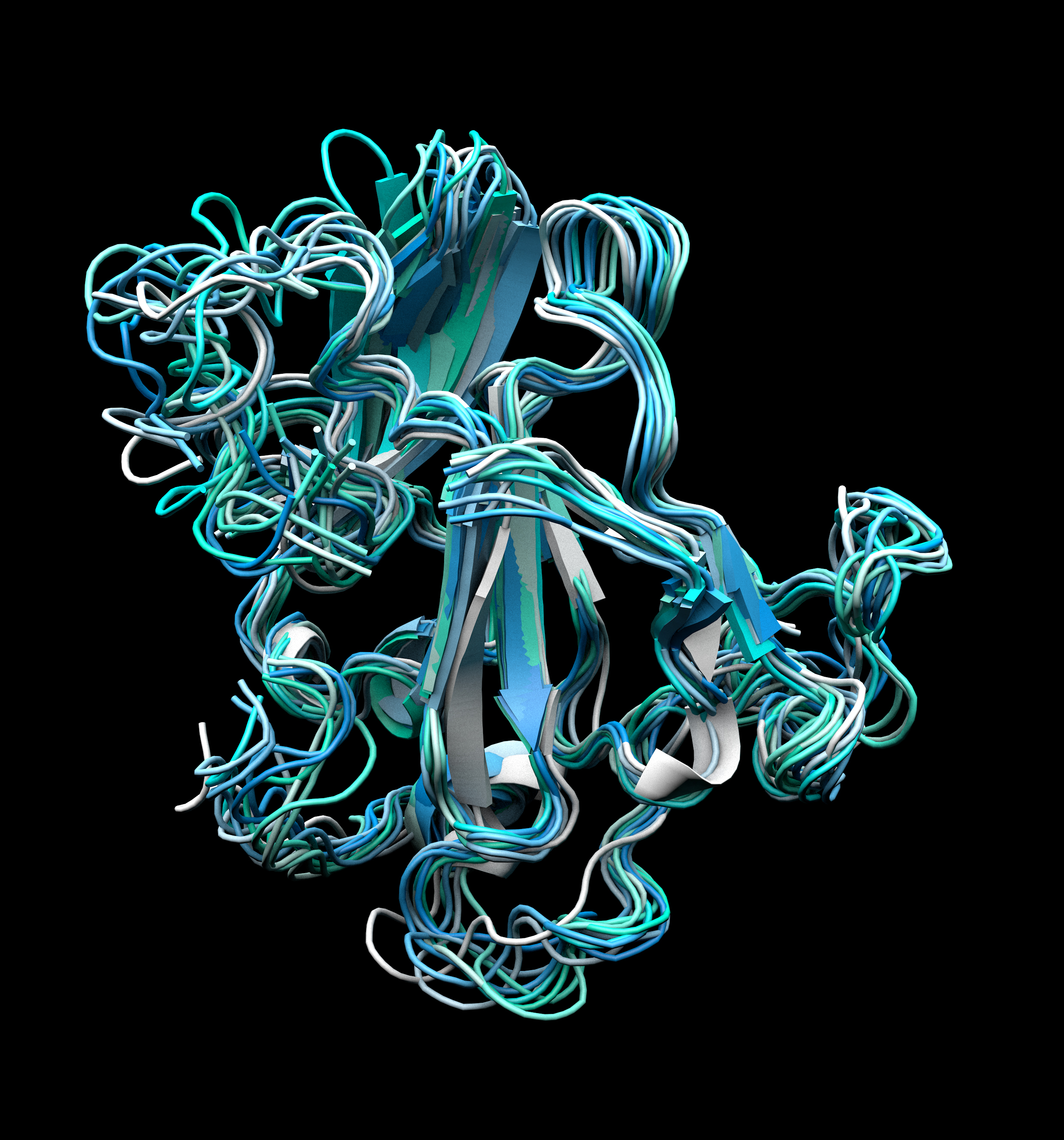
To understand why, the Law and Wilson laboratories teamed up with TSRI Associate Professor Andrew Ward and used electron microscopy and several other advanced structural analysis tools to take a closer look at HCV’s E2 protein, in particular the dynamics of its receptor binding site. Their investigations focused on the “recombinant” form of the E2 protein, produced in the lab and therefore isolated from the rest of the virus. Recombinant E2 is a prime candidate for HCV vaccine design and is much easier to purify and study than E2 from whole virus particles.
One finding was that recombinant E2, probably due to its many strong disulfide bonds, has great structural stability, with an unusually high melting point of 85°C. However, the TSRI scientists also found evidence that, within this highly buttressed construction, the receptor binding site portion is extraordinarily loose and flexible in the recombinant protein.
“It adopts a very wide range of conformations,” said study first author Leopold Kong, of TSRI at the time of the study, now at the National Institutes of Health.
Prior studies have shown that HCV’s receptor binding site adopts a narrow range of conformations (shapes) when bound by virus-neutralizing antibodies. A vaccine that elicited high levels of antibodies against only these key conformations would in principle provide effective protection. But this study suggests that the E2 protein used in candidate vaccines displays far too many other binding-site conformations—and thus elicits antibodies that mostly do nothing to stop the actual virus.
Law and Wilson and their colleagues plan to follow up by studying E2 and its receptor binding site as they are presented on the surface of the actual virus. They also plan to design a new version of E2 or even an entirely different scaffold protein, on which the receptor binding site is stabilized in conformations that will elicit virus-neutralizing antibodies.
In addition to Law, Wilson, Ward and Kong, authors of the study, “Structural flexibility at a major conserved antibody target on hepatitis C virus E2 antigen,” included Rameshwar U. Kadam, Erick Giang, Travis Nieusma, Fernando Garces and Netanel Tzarum, all of TSRI; and David E. Lee, Tong Liu, Virgil Woods and Sheng Li, of the University of California, San Diego.
Support for the study was provided by the National Institutes of Health (AI079031, AI123861, AI106005, AI123365, GM094586, AI117905, GM020501 and AI101436) and TSRI’s Skaggs Institute for Chemical Biology.
For more information, contact press@scripps.edu See More News

