The Boddy Laboratory
Research
Guardians of Genome Stability
Loss of genome stability is causally associated with a number of human diseases including cancer, neurological and developmental disorders. Safeguards that promote faithful passage of the genome include cell cycle checkpoints and DNA repair mechanisms. DNA integrity checkpoints both prevent commitment to mitosis in the presence of unreplicated or damaged DNA and contribute to coordinating the appropriate DNA repair responses. We study critical cellular processes at the interface of cell cycle checkpoints and DNA repair, which has uncovered key insights on leukemia therapy and promises to reveal other disease mechanisms and potential therapeutic targets.
An Unprecedented SUMO-Targeted Ubiquitin Ligase (STUbL)
The covalent attachment of ubiquitin and SUMO to target proteins plays key roles in genome stability; with each moiety having physiologically distinct effects on target protein function. We have identified the SUMO-Targeted Ubiquitin Ligase (STUbL) family, which provides a novel and unanticipated regulatory link between the ubiquitination and sumoylation pathways. The STUbL family includes the founding members: fission yeast Slx8-Rfp1, human RNF4, slime mold MIP1 and budding yeast SLX5/8. STUbLs are recruited to sumoylated proteins and proteins containing SUMO-like domains, to mediate their ubiquitination and degradation (Figure 1). Cells with mutations in Slx8-Rfp1 accumulate sumoylated proteins, display genomic instability and are hypersensitive to genotoxic stress. These Slx8-Rfp1 mutant phenotypes are suppressed by concomitant deletion of the major SUMO ligase Pli1, demonstrating the specificity of STUbLs as regulators of sumoylated proteins. Expression of human RNF4 restores SUMO pathway homeostasis in fission yeast that lack Slx8-Rfp1, underscoring the evolutionary functional conservation of STUbLs. The DNA repair factor Rad60 (an accessory factor of the Smc5-6 complex) and its human homologue NIP45, which contain SUMO-like domains, are candidate STUbL targets. Consistently, mutations in Rad60 and Slx8-Rfp cause similar DNA repair defects.
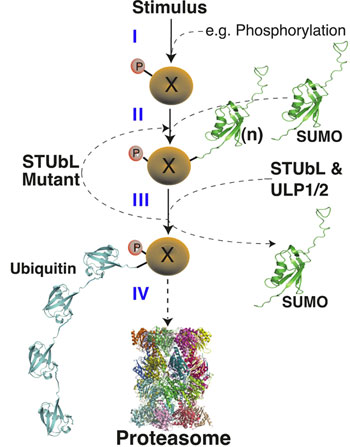
Figure 1. STUbL Pathway. (I-II) After stimulus, target X becomes sumoylation competent (e.g. phosphorylated) and is SUMO conjugated. (III) Following execution of the SUMO conjugated target function, STUbL-mediated ubiquitination, perhaps coupled to desumoylation by Ulp1/2, promotes target proteasomal target degradation (IV). SUMO recycling may also occur at the proteasome as for ubiquitin.
Discovery of SUMO Mimicry, Key to Genome Stability
Posttranslational modification of proteins by the Small Ubiquitin-like MOdifier SUMO has emerged as a critical regulator of genome stability. SUMO is covalently attached to target proteins via an enzymatic cascade, consisting of E1 activating, E2 conjugating and E3 ligase factors. SUMO attachment exerts multiple regulatory effects on target proteins including altering their localization, interactions, conformation and stability. In collaboration with John Tainer’s laboratory (TSRI), we identified unanticipated regulatory interplay between the DNA repair protein Rad60, and the enzymes required for posttranslational protein modification by SUMO. Rad60 is an intriguing protein that contains integral tandem globular domains with weak similarity to SUMO, which we call SUMO-like domains (SLD1 and 2). The ultra-high resolution (0.97Å) structure of Rad60 SLD2 revealed several important features (Figure 2). SLD2, like SUMO, adopts the b-grasp fold common to the ubiquitin-like protein superfamily. Most crucially, the structure revealed that the surface features of SUMO and SLD2 are quite distinct, with the interesting exception of a small conserved area required for non-covalent interaction between SUMO and its E2 conjugating enzyme Ubc9 (Figure 2). Consistently, we found that SLD2 interacts with Ubc9, but not the other non-covalent SUMO interactors. Using structure-guided mutational analyses we specifically uncoupled the Rad60 SLD2:Ubc9 interface to determine its physiological significance. Abrogating the SLD2:Ubc9 interface in vivo resulted in spontaneous DNA damage, genomic instability and hypersensitivity to genotoxic stresses. Further genetic analyses indicated a role for the SLD2:Ubc9 interface in suppressing pathological replication fork-associated recombination processes. The intimate functional interplay between the SUMO pathway and Rad60 provides the first example of molecular mimicry in the SUMO pathway. Given the pivotal role played by SUMO modification in genome stabilization and in certain disease processes, it will be important to determine the precise role of Rad60 in the SUMO pathway.
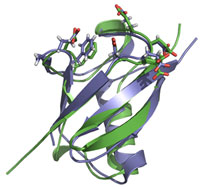
Figure 2. Discovery of SUMO Mimicry and its Role in Genome Stability. The image depicts the backbone structures of SUMO in green superimposed on the second Rad60 SUMO-like domain (SLD2) in blue. Residues at the conserved interface for binding to the SUMO E2 enzyme are highlighted in both molecules. This surface of Rad60 is critical for its mimicry of SUMO and for its roles in genome stability. Notably, other residues in SUMO used for additional critical interactions are not conserved in Rad60 SLD2.
Sites and Mechanisms of Smc5-6 Loading on Chromosomes
The octameric structural maintenance of chromosomes (SMC) Smc5-6 complex plays essential roles in chromosome segregation and DNA repair (Figure 3). Smc5-6 mutant cells suffer catastrophic chromosome segregation defects, in part due to improper engagement/completion of the homologous recombination DNA repair pathway. In collaboration with the laboratory of Steven Head (TSRI) we have identified “hot spots” of Smc5-6 activity by defining the sites at which the complex is enriched on chromosomes. We determined that Smc5-6 loads at the heterochromatic centromeres of the three fission yeast chromosomes and plays a role in the faithful segregation of these repetitive and recombination-prone loci (Figure 3; Clr4 histone methyltransferase-dependent). Intriguingly, in response to DNA damage, Smc5-6 loads at subtelomeric regions of chromosomes in a manner dependent on posttranslational modification of as yet unknown targets by SUMO, via the Smc5-6 E3 SUMO ligase Nse2 (Figure 3). Finally, we found that Smc5-6 loads at all transcriptionally active chromosomal transfer RNA encoding genes (tDNAs). Overall, the nature of the sites at which Smc5-6 is enriched supports a role for Smc5-6 in suppressing illegitimate recombination or gross chromosomal rearrangements (GCRs) at these loci. GCRs are common priming events in human disease, including cancer.
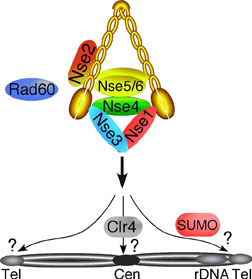
Figure 3. Sites and Mechanisms of Smc5-6 Loading on Chromosomes. We have compositionally defined the octameric Smc5-6 genome stability holocomplex and have now determined, genome-wide, the sites and mechanisms of Smc5-6 chromosome loading. Tel, indicates telomeric sites; Cen, heterochromatic centromeric loci and rDNA, the ribosomal DNA loci. SUMO is required for telomeric recruitment and Clr4 (histone methyltransferase) for the centromeric localization of Smc5-6.