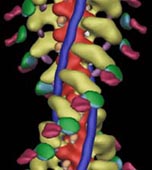
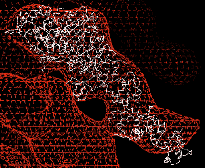
 Three-dimensional (3D) maps of decorated filaments calculated by cryo-EM and image analysis at
a resolution of 25-30 Å show the overall shape of the myosin head and its geometry of attachment
to actin. Light chains and small discrete domains can be localized in three dimensions by difference mapping,
and undecagold labeling of specific amino acid residues coupled with difference mapping can be used to locate
regions of the polypeptide chain accurately (Fig 1). Such data provide strong
constraints for docking the X-ray structures of actin and the myosin head into the 30 Å maps of the
actomyosin complex (Fig. 2) (Rayment et al, 1993). This approach allows
identification of the regions of actin and myosin that interact. However, it does not provide a detailed
picture of the interacting surfaces, as the model of this strongly bound state is built from the structure
of a weak binding myosin conformation. Visualizing the details of the interacting surfaces remains one of
the challenges for the future.
Three-dimensional (3D) maps of decorated filaments calculated by cryo-EM and image analysis at
a resolution of 25-30 Å show the overall shape of the myosin head and its geometry of attachment
to actin. Light chains and small discrete domains can be localized in three dimensions by difference mapping,
and undecagold labeling of specific amino acid residues coupled with difference mapping can be used to locate
regions of the polypeptide chain accurately (Fig 1). Such data provide strong
constraints for docking the X-ray structures of actin and the myosin head into the 30 Å maps of the
actomyosin complex (Fig. 2) (Rayment et al, 1993). This approach allows
identification of the regions of actin and myosin that interact. However, it does not provide a detailed
picture of the interacting surfaces, as the model of this strongly bound state is built from the structure
of a weak binding myosin conformation. Visualizing the details of the interacting surfaces remains one of
the challenges for the future.
| The use of cryo-EM also provided the first views of large scale conformational changes in the actomyosin complex in response to nucleotide. A comparison of 3D maps of, for example, actin decorated with smooth muscle myosin II heads in the presence and absence of Mg-ADP shows that the light chain-containing domain swings 23° in response to nucleotide release (Fig. 3) (Whittaker et al, 1995). Subsequently, a number of other myosins (e.g., brush border myosin I, myosin VI) have been shown to exhibit similar nucleotide-dependent behavior (Jontes et al, 1995, Wells et al 1999). It appears that in some, but not all, myosins, the ADP release step is associated with an additional swing of the light chain domain 'powerstroke'. Whether this step produces force or whether it functions as a movement sensor in myosins that work cooperatively to achieve processivity with only a few motor molecules remains to be seen. | 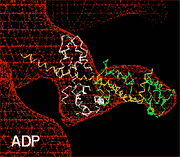 |
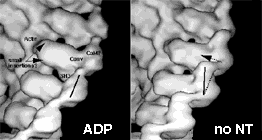 |
One fascinating result from the work on nucleotide-induced conformational changes was the observation that class VI myosins swing their light chain domain in the opposite direction to other myosins on ADP release (Fig. 4) (Wells et al, 1999). Indeed, these myosins move 'backwards' on actin filaments. Their role in the cell is currently being elucidated in the Kendrick-Jones and Hasson labs. |
References
Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC & Milligan RA 1993 Structure of the actin-myosin complex and its implications for muscle contraction. Science 261, 58-65
Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA & Sweeney HL
1999 Myosin VI is an actin-based motor that moves backwards. Nature 401, 505-508
Whittaker M, Wilson-Kubalek EM, Smith JE, Faust L, Milligan RA & Sweeney HL 1995 A 35-Å movement of smooth muscle myosin on ADP release. Nature 378, 748-751
View links to recent papers on myosin structure from PUBMED
Return to Milligan Lab Home Page
Jontes JD, Wilson-Kubalek EM & Milligan RA
1995 A 32 degree tail swing in brush border myosin I on ADP release. Nature 378, 751-753
Return to Milligan Lab Research Page
contents © 2000
awl