- Romesberg Lab
- Research
- Publications

Romesberg Lab Publications
Browse it all on PubMed or browse here by topic:
Genetic Alphabet Biophysics Combating Bacteria Reviews
Browse it all on PubMed or browse here by topic:
Genetic Alphabet Biophysics Combating Bacteria ReviewsOngoing since 1998, the lab has worked to develop unnatural base pairs (UBPs), achieving over the years stability in duplex DNA, acceptance by native polymerases, compatibility with PCR, linker modifications to allow attachment of novel functionality, and storage of information in a living cell. Related work included the evolution of polymerases to accept the UBPs, which more recently has transitioned to application of the mutant polymerases for SELEX to evolve stable aptamers made up of modified or unnatural nucleotides.
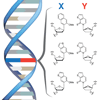
We report a systematic extension of the effort to optimize the SSO by exploring a variety of deoxy- and ribonucleotide analogues, and report the first in vivo structure–activity relationship (SAR) analysis of unnatural ribonucleoside triphosphates.
AW Feldman, VT Dien, RJ Karadeema, EC Fischer, Y You, BA Anderson, R Krishnamurthy, JS Chen, L Li, FE Romesberg (2019) J Am Chem Soc, published 26 June
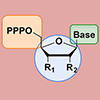
We synthesize two novel benzothiophene-based dNaM analogs, dPTMO and dMTMO, and characterize the corresponding UBPs with dTPT3. We demonstrate that these UBPs perform similarly to, or slightly worse than, dNaM-dTPT3 in vitro; but in the in vivo environment of a semi-synthetic organism, retention of dMTMO-dTPT3 and dPTMO-dTPT3 are significantly higher. The results demonstrate the importance of evaluating synthetic biology “parts” in their in vivo context
VT Dien, M Holcomb, AW Feldman, EC Fischer, TJ Dwyer, FE Romesberg (2018) J Am Chem Soc 140:16115–16123
We characterize the effects of nucleobase, sugar, and triphosphate modifications on the import of nucleoside triphosphates into E. coli by the transporter PtNTT2 using a variety of assays. Overall, our results demonstrate the general utility of PtNTT2-mediated import of natural or modified nucleoside triphosphates for different applications in molecular or synthetic biology.
AW Feldman, EC Fischer, MP Ledbetter, J-Y Liao, JC Chaput, FE Romesberg (2018) J Am Chem Soc 140:1447–1454

We explore the contributions of the E. coli DNA replication and repair machinery to the propagation of DNA containing dNaM-dTPT3 and show that replication by DNA polymerase III, supplemented with the activity of polymerase II and methyl-directed mismatch repair contribute to retention of the UBP and that recombinational repair of stalled forks is responsible for the majority of its loss.
MP Ledbetter, RJ Karadeema, FE Romesberg (2018) J Am Chem Soc 140:758-765
We report the in vivo transcription of DNA containing dNaM and dTPT3 into mRNAs with two different unnatural codons and tRNAs with cognate unnatural anticodons, and their efficient decoding at the ribosome to direct the site-specific incorporation of natural or non-canonical amino acids into superfolder green fluorescent protein.
Y Zhang, JL Ptacin, EC Fischer, HR Aerni, CE Caffaro, K San Jose, AW Feldman, CR Turner, FE Romesberg (2017) Nature 551:644-647
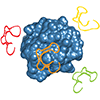
We report the first selection of fully 2′-OMe modified aptamers, specifically aptamers that bind human neutrophil elastase (HNE). Six aptamers are shown to bind with reasonable affinity, and affinity requires the 2′-OMe substituents. A complete characterization of one of the aptamers demonstrates that affinity for HNE is retained in the presence of high concentrations of salt or serum.
Z Liu, T Chen, FE Romesberg (2017) Chem Sci 8:8179-8182

We report that the polymerase mutant SFM4-3 can efficiently synthesize polymers composed of nucleotides with 2′-azido, 2′-chloro, 2′-amino, or arabinose sugars, and that SFM4-3 can PCR amplify these modified oligonucleotides. Amplified polymers may be functionalized with desired moieties arrayed in a controlled fashion. Using this approach, we produce a novel DNA hydrogel. Read and hear more in The San Diego Union Tribune.
T Chen, FE Romesberg (2017) Angew Chem Int Ed 56:14046–14051
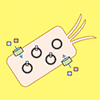
We screen 135 candidate unnatural base pairs (UBPs) for optimal performance in an E. coli semi-synthetic organism (SSO). Interestingly, we find that in vivo SARs differ from those collected in vitro, and most importantly, we identify four UBPs whose retention in the DNA of the SSO is higher than that of dNaM-dTPT3.
AW Feldman, FE Romesberg (2017) J Am Chem Soc 139:11427–11433

We reply to a letter that suggests that our hydrophobic UBPs are not suitable for the expansion of the genetic alphabet.
AW Feldman, MP Ledbetter, Y Zhang, FE Romesberg (2017) Proc Natl Acad Sci USA 114:E6478–E6479
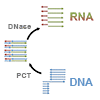
We show that the evolved Stoffel fragment mutant, SFM4-3, efficiently transcribes RNA or 2′-F-modified RNA and that it also efficiently PCR amplifies oligonucleotides of mixed RNA and DNA composition. Using thermocycling and a novel DNA template, we demonstrate a polymerase chain transcription (PCT) reaction that results in the exponential production of orders of magnitude more RNA or modified RNA than is available by conventional transcription.
T Chen, FE Romesberg (2017) J Am Chem Soc 139:9949–9954
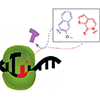
We report a steady-state kinetic characterization of the rate with which the Klenow fragment of E. coli DNA polymerase I synthesizes the dNaM-dTPT3 UBP and its mispairs in a variety of sequence contexts. The data demonstrate that dNaMTP and dTPT3TP are well optimized and standardized parts for the expansion of the genetic alphabet.
SE Morris, AW Feldman, FE Romesberg (2017) ACS Synth Biol 6:1834–1840
We use the thermostable DNA polymerase, SFM4-3, in selections for 2′-F purine aptamers that bind human neutrophil elastase (HNE). We show that inclusion of only a few 2′-F substituents can optimize aptamer properties far beyond simple nuclease resistance and that SFM4-3 should prove valuable for the further exploration and production of aptamers with properties optimized for various applications.
D Thirunavukarasu, T Chen, Z Liu, N Hongdilokkul, FE Romesberg (2017) J Am Chem Soc 139:2892-2895
We report the synthesis of unnatural triphosphates with their β,γ-bridging oxygen replaced with a difluoromethylene moiety, yielding dNaMTPCF2 and dTPT3TPCF2, and evaluate their retention in an E. coli SSO. Correction: Note that in the Degradation of Nucleotide Triphosphates section of the Methods, cells were propagated in media containing 200 µM nucleotide triphosphate, not 200 mM.
AW Feldman, FE Romesberg (2017) J Am Chem Soc 139:2464-2467
We describe an optimized SSO that is healthy, more autonomous than its predecessor, and able to store increased information indefinitely. The SSO constitutes a stable form of semisynthetic life and lays the foundation for efforts to impart life with new forms and functions. Read more in The Guardian, The Washington Post, TSRI News Release, and The San Diego Union Tribune, or listen to an interview in This Way Up from Radio New Zealand.
Y Zhang, BM Lamb, AW Feldman, AX Zhou, T Lavergne, L Li, FE Romesberg (2017) Proc Natl Acad Sci USA 114:1317–1322
We report the development of a polymerase evolution system and its use to evolve thermostable polymerases that efficiently interconvert C2'-OMe modified oligonucleotides and their DNA counterparts via “transcription” and “reverse transcription,” or more importantly, PCR amplify partially C2'-OMe or C2'-F modified oligonucleotides.
T Chen, N Hongdilokkul, Z Liu, R Adhikary, SS Tsuen, FE Romesberg (2016) Nat Chem 8:556-562
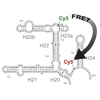
We report the synthesis and evaluation of several unnatural ribotriphosphates bearing linkers that allow the chemoselective attachment of different functionalities. One unnatural base pair is used to dual label a 243-nt fragment of a 16S RNA with Cy3 and Cy5, which are then used to characterize conformational changes in the presence of ribosomal proteins.
T Lavergne, R Lamichhane, DA Malyshev, Z Li, L. Li, E Sperling, JR Williamson, DP Millar, FE Romesberg (2016) ACS Chem Biol 11:1347-1353
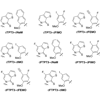
We report a PCR-based screen to identify promising unnatural base pairs from a library of 111 unnatural nucleotides (~6,000 candidate base pairs).
K Dhami, DA Malyshev, P Ordoukhanian, T Kubelka, M Hocek, FE Romesberg (2014) Nucleic Acids Res 42:10235-10244
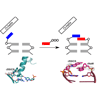
We demonstrate that E. coli can stably maintain a plasmid containing d5SICS–dNaM with only the addition of an exogenously expressed algal nucleotide triphosphate transporter. Read the associated Nature News & Views or the article in the New York Times
DA Malyshev, K Dhami, T Lavergne, T Chen, N Dai, JM Foster, IR Corrêa, FE Romesberg (2014) Nature 509:385-388

In this work, we identify dTPT3-dNaM, which is PCR amplified with a natural base pair-like efficiency and fidelity. We also show that the dTPT3 scaffold is uniquely tolerant of attaching a propargyl amine linker, enabling site-specific labeling of PCR-amplified DNA.
L Li, M Degardin, T Lavergne, D Malyshev, K Dhami, P Ordoukhanian, FE Romesberg (2014) J Am Chem Soc 136:826-829
We report the structural characterization of the prechemistry complexes with KlenTaq polymerase corresponding to the insertion of dNaMTP opposite d5SICS, as well as mulitple postchemistry complexes in which the already formed unnatural base pair is positioned at the postinsertion site.
K Betz, DA Malyshev, T Lavergne, W Welte, K Diederichs, FE Romesberg, A Marx (2013) J Am Chem Soc 135:18637-18643
We report the synthesis and characterization of unnatural base pairs bearing propynyl groups, and their use to site-specifically label amplified DNA via Click chemistry. We demonstrate the attachment of a small molecule biotin tag, and for the first time a protein (SH3), to a large, PCR amplified fragment of DNA, as well as the arraying of two proteins on the same duplex.
Z Li, T Lavergne, DA Malyshev, J Zimmermann, R Adhikary, K Dhami, P Ordoukhanian, Z Sun, J Xiang, FE Romesberg (2013) Chem Eur J 19:14205-14209
Here, we report the stepwise optimization of dMMO2 via the synthesis and evaluation of eighteen novel para-derivatized analogs of dMMO2, followed by further derivatization and evaluation of the most promising analogs with meta substituents. The most promising, d5SICS-dFEMO, is replicated under some conditions with greater efficiency and fidelity than d5SICS-dNaM.
T Lavergne, M Degardin, DA Malyshev, HT Quach, K Dhami, P Ordoukhanian, FE Romesberg (2013) J Am Chem Soc 135:5408-5419
We examine the PCR amplification of DNA containing one or more d5SICS-dNaM pairs and demonstrate that this DNA may be amplified with high efficiency and with greater than 99.9% fidelity. The results demonstrate that for PCR and PCR-based applications, d5SICS-ddNaM is functionally equivalent to a natural base pair, and when combined with dA-dT and dG-dC, provides the first fully functional six-letter genetic alphabet.
DA Malyshev, K Dhami, HT Quach, T Lavergne, P Ordoukhanian, A Torkmani, FE Romesberg (2012) Proc Natl Acad Sci USA 109:12005-12010
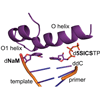
We report crystal structures of KlenTaq DNA polymerase at different stages of replicating dNaM-d5SICS, and show that efficient replication results from the polymerase itself inducing the required natural-like structure.
K Betz, DA Malyshev, T Lavergne, W Welte, K Diederichs, TJ Dwyer, P Ordouklanian, FE Romesberg, A Marx (2012) Nat Chem Biol 8:612-614
The continued optimization of a predominantly hydrophobic class of unnatural base pairs is reported. We find that minimizing the cost of nucleobase desolvation and optimizing packing interactions within the developing major groove of DNA is promising route toward optimization. While we identify an improved base pair, dNMO1-d5SICS, continued analysis of the previously reported dNaM-d5SICS pair reveals that it is replicated with an efficiency and fidelity approaching those of natural DNA.
T Lavergne, DA Malyshev, FE Romesberg (2012) Chem Eur J 18:1231-1239
We report the synthesis and analysis of the ribo- and deoxyribo-variants, (d)5SICS and (d)MMO2, modified with free or protected propargylamine linkers that allow for the site-specific modification of DNA or RNA during or after enzymatic synthesis. We also synthesized and evaluated the α-phosphorothioate variant of d5SICSTP, which provides a route to backbone thiolation and an additional strategy for the post-amplification site-specific labeling of DNA.
YJ Seo, DA Malyshev, T Lavergne, P Ordoukhanian, FE Romesberg (2011) J Am Chem Soc 133:19878-19888
Here, we synthesize a variety of derivatized unnatural base pairs and characterize their structures and DNA polymerase-mediated replication. We also present further evidence supporting an intercalation-based mechanism of replication, and show that dDMO—d5SICS represents significant progress toward the expansion of the genetic alphabet.
DA Malyshev, DA Pfaff, SI Ippoliti, GT Hwang, TJ Dwyer, FE Romesberg (2010) Chem Eur J 16:12650-12659
We identified a mutant of Taq DNA polymerase that incorporates fluorophore-labeled substrates 50 to 400-fold more efficiently into scarred primers in solution and also demonstrates significantly improved performance under actual sequencing conditions.
AM Leconte, MP Patel, LE Sass, P McInerney, M Jarosz, L Kung, JL Bowers, PR Buzby, JW Efcavitch, FE Romesberg (2010) Angew Chem Int Ed 34:5921-5924

We demonstrate that DNA containing either the d5SICS−dNaM or d55SICS−dMMO2 unnatural base pair may be amplified by PCR with fidelities and efficiencies that approach those of fully natural DNA.
DA Malyshev, YJ Seo, P Ordoukhanian, FE Romesberg (2009) J Am Chem Soc 131:14620-14621

Cover Story: We synthesize and evaluate several dMMO2 derivatives with meta-chlorine, -bromine, -iodine, -methyl, or -propinyl substituents. Complete characterization of unnatural base pair and mispair synthesis and extension reveal that the modifications have large effects only on the efficiency of unnatural base pair synthesis and that the effects likely result from a combination of changes in steric interactions, polarity, and polarizability.
YJ Seo, FE Romesberg (2009) ChemBioChem 10:2394-2400
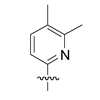
We examine six pyridine-based nucleotides, differentiated by methyl substitution, that are designed to vary both inter- and intra-strand packing within duplex DNA. We show that incorporation of one of these nucleobases, d34DMPy, as a self pair or as a mispair with dA significantly increases oligonucleotide hybridization fidelity at other positions within the duplex.
GT Hwang, Y Hari, FE Romesberg (2009) Nucleic Acids Res 37:4757-4763
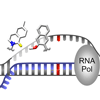
We show here that both of the unnatural base pairs d5SICS:dMMO2 and d5SICS:dNaM are selectively transcribed by T7 RNA polymerase and that the efficiency of d5SICS:dNaM transcription in both possible strand contexts is only marginally reduced relative to that of a natural base pair.
YJ Seo, S Matsuda, FE Romesberg (2009) J Am Chem Soc 131:5046-5047
To better understand and optimize the slowest step of replication of the unnatural base pair formed between the nucleotides dMMO2 and d5SICS, the insertion of MMO2 opposite d5SICS, we synthesize two dMMO2 derivatives, d5FM and dNaM, which differ from the parent nucleobase in terms of shape, hydrophobicity, and polarizability, and we characterize their enzymatic replication.
YJ Seo, GT Hwang, P Ordoukhanian, FE Romesberg (2009) J Am Chem Soc 131:3246-3252

We examine the ability of five different polymerases (family A polymerases from bacteriophage T7 and Thermus aquaticus, family B polymerases from Thermococcus litoralis and Thermococcus 9°N-7, and the family X polymerase, human polymerase β.) to replicate DNA containing four different unnatural nucleotides bearing predominantly hydrophobic nucleobase analogs, and we identify some aspects of recognition that are conserved.
GT Hwang, FE Romesberg (2008) J Am Chem Soc 130:14872-14882
We demonstrate that the pyridyl nucleobase scaffold can be optimized for replication, both as a self pair and as a component of a heteropair, and we identify a pyridyl-nucleotide self pair that is well recognized by a DNA polymerase, allowing it to be replicated and used to synthesize site-specifically labeled DNA in good yields.
Y Hari, GT Hwang, AM Leconte, N Joubert, M Hocek, FE Romesberg (2008) ChemBioChem 9:2796-2799
Read about this article at New Scientist, JAMA, or Popular Science.
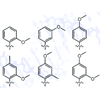
AM Leconte, GT Hwang, S Matsuda, P Capek, Y Hari, FE Romesberg (2008) J Am Chem Soc 130:2336-2343
We synthesize and characterize three fluoro-substituted pyridone nucleoside analogs. Generally, we find that the specific fluorine substitution pattern of the analogs has little impact on unnatural pair or mispair stability, but does significantly increase the rate at which the pyridone-based unnatural base pairs are extended.
GT Hwang, AM Leconte, FE Romesberg (2007) ChemBioChem 8:1606-1611

We examine the structure of DNA duplexes containing either the d3FB or dPICS self-pairs. We find that the large aromatic rings of the dPICS nucleobases pair in an intercalative manner within duplex DNA, while the d3FB nucleobases interact in an edge-on manner, much closer in structure to natural base pairs.
S Matsuda, JD Fillo, AA Henry, P Rai, SJ Wilkens, TJ Dwyer, BH Geierstanger, DE Wemmer, PG Schultz, G Spraggon, FE Romesberg (2007) J Am Chem Soc 129:10466-10473
We examine the enzymatic synthesis of DNA with six different unnatural nucleotides bearing methoxy-derivatized nucleobase analogues. Different nucleobase substitution patterns were used to systematically alter the nucleobase electronics, sterics, and hydrogen-bonding potential. Our results indicate that ortho methoxy groups should be generally useful for the effort to expand the genetic alphabet.
S Matsuda, AM Leconte, FE Romesberg (2007) J Am Chem Soc 129:5551-5557
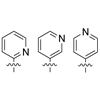
Pyridyl nucleoside analogues are synthesized and characterized (see structures). An α-glycosidic nitrogen atom provides an H-bond acceptor that does not significantly facilitate pairing with natural nucleobases. However, it forms minor-groove H-bonds with water molecules and DNA polymerases that optimize the stability and replication, respectively, of the unnatural base pair.
Y Kim, AM Leconte, Y Hari, FE Romesberg (2006) Angew Chem Int Ed 45:7809-7812
Here, we report the synthesis and characterization of nucleotides bearing pyridone nucleobase analogues, which are expected to position their carbonyl group into the minor groove.
AM Leconte, S Matsuda, GT Hwang, FE Romesberg (2006) Angew Chem Int Ed 45:4326-4329
We describe a simple screen that enables the characterization of large numbers of previously uncharacterized base pairs. From this screen, we identify a class of unnatural base pairs which are extended more efficiently than any unnatural base pair reported to date.
AM Leconte, S Matsuda, FE Romesberg (2006) J Am Chem Soc 128:6780-6781
We report the characterization of polymerase-mediated replication of unnatural base pairs formed between nucleotides bearing simple methyl-substituted phenyl ring nucleobase analogues.
S Matsuda, AA Henry, FE Romesberg (2006) J Am Chem Soc 128:6369-6375
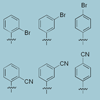
We report the synthesis, stability and polymerase recognition of nucleoside analogs bearing single bromo- or cyano-derivatized phenyl rings and find that both modifications generally stabilize base pair formation to a greater extent than methyl or fluoro substitution.
GT Hwang, FE Romesberg (2006) Nucleic Acids Res 34:2037-2045
We report the use of an activity-based selection system to evolve a DNA polymerase that more efficiently replicates DNA containing the PICS self-pair.
AM Leconte, L Chen, FE Romesberg (2005) J Am Chem Soc 127:12470-12471
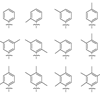
We report the synthesis and stability of unnatural base pairs formed between simple phenyl rings modified at different positions with methyl groups. Several are virtually as stable as a natural base pair in the same sequence context, showing that neither hydrogen-bonding nor large aromatic surface area are required for base pair stability within duplex DNA and that interstrand interactions between small aromatic rings may be optimized for both stability and selectivity.
S Matsuda, FE Romesberg (2004) J Am Chem Soc 126:14419-14427
Read about this article in The Econimist or Radio Canada.
AA Henry, AG Olsen, S Matsuda, C Yu, BH Geierstanger, FE Romesberg (2004) J Am Chem Soc 126:6923-6931

In this report, mutants that efficiently synthesize an unnatural polymer from 2‘-O-methyl ribonucleoside triphosphates were immobilized and isolated by means of an activity-dependent phage display method. One evolved polymerase is shown to incorporate the modified substrates with an efficiency and fidelity equivalent to that of the wild-type enzyme with natural substrates.
M Fa, A Radeghieri, AA Henry, FE Romesberg (2004) J Am Chem Soc 126:1748-1754
We report the stability and replication of DNA containing self-pairs formed between unnatural nucleotides bearing benzofuran, benzothiophene, indole, and benzotriazole nucleobases.
S Matsuda, AA Henry, PG Schultz, FE Romesberg (2003) J Am Chem Soc 125:6134-6139
We report the stability and replication of DNA containing self-pairs formed between unnatural nucleotides differing in their substitution with oxygen and sulfur atoms.
AA Henry, C Yu, FE Romesberg (2003) J Am Chem Soc 125:9638-9646
We report our initial efforts toward the development of an unnatural in vivo nucleoside phosphorylation pathway that is based on nucleoside salvage enzymes.
Y Wu, M Fa, EL Tae, PG Schultz, FE Romesberg (2002) J Am Chem Soc 124:14626-14630
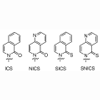
Heteroatom substitution of ICS via replacement of C6 with nitrogen and thio substitution at C10 provides the base SNICS, which forms stable self-pairs and can therefore be used for efficient unnatural base pair replication.
C Yu, AA Henry, FE Romesberg, PG Schultz (2002) Angew Chem Int Ed 41:3841-3844

We use an activity-based phage display method to screen a Stoffel fragment (SF) polymerase library, and we identify three SF mutants that incorporate ribonucleoside triphosphates virtually as efficiently as the wild-type SF polymerase incorporates dNTP substrates.
G Xia, L Chen, T Sera, M Fa, PG Schultz, FE Romesberg (2002) Proc Natl Acad Sci USA 99:6597-6602
Here, we examine thiophene and furan heterocycles as nucleobase analogues. The stability of the unnatural bases was evaluated in duplex DNA paired opposite other unnatural bases as well as opposite the natural bases. Several unnatural base pairs are identified that are both reasonably stable and strongly selective against mispairing with native bases.
M Berger, SD Luzzi, AA Henry, FE Romesberg (2002) J Am Chem Soc 124:1222-1226

We show that the unnatural self-pair formed between two 7-azaindole nucleosides can be replicated in DNA using binary polymerase system - KF efficiently synthesizes the self-pair, and pol β efficiently extends it.
EL Tae, Y Wu, G Xia, PG Schultz, FE Romesberg (2001) J Am Chem Soc 123:7439-7440
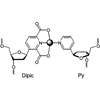
We describe a base pair with a pyridine-2,6-dicarboxylate nucleobase (Dipic) as a planar tridentate ligand, and a pyridine nucleobase (Py) as the complementary single donor ligand. Characterization of this base pair in duplex DNA shows that interbase metal coordination can replace the hydrogen bonding schemes found in the natural base pairs dA:dT and dG:dC.
E Meggers, PL Holland, WB Tolman, FE Romesberg, PG Schultz (2000) J Am Chem Soc 122:10714-10715
Here we report the synthesis and characterization of 3MN and 2Np, and we show that the 3MN:3MN self-pair is synthesized by KF with an efficiency of 2.4 × 106 M-1min-1.
AK Ogawa, Y Wu, M Berger, PG Schultz, FE Romesberg (2000) J Am Chem Soc 122:8803-8804
Relying on only interbase hydrophobic packing for bonding, several unnatural nucleobases are reported that form selective and stable self-pairs in duplex DNA. These unnatural nucleobases should make it possible to increase the stringency and information content of hybridization or encoding experiments.
M Berger, AK Ogawa, DL McMinn, Y Wu, PG Schultz, FE Romesberg (2000) Angew Chem Int Ed 39:2940-2942
Read about this article in Science or The New York Times.
Y Wu, AK Ogawa, M Berger, DL McMinn, PG Schultz, FE Romesberg (2000) J Am Chem Soc 122:7621-7632
AK Ogawa, Y Wu, DL McMinn, J Liu, PG Schultz, FE Romesberg (2000) J Am Chem Soc 122:3274-3287

The reported unnatural, predominantly hydrophobic nucleoside analogs are efficient ‘universal bases’ for hybridization, template directed DNA synthesis and chain termination. Moreover, several of the universal bases function in their biological role, hybridization or replication, with an efficiency not significantly reduced relative to their natural counterparts.
M Berger, Y Wu, AK Ogawa, DL McMinn, PG Schultz, FE Romesberg (2000) Nucleic Acids Res 28:2911–2914
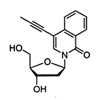
This study demonstrates that hydrophobicity is a sufficient driving force for the stable and selective pairing of the 7-propynyl isocarbostyril nucleoside in duplex DNA, as well as for the selective enzymatic incorporation of this nucleobase against itself during DNA synthesis.
DL McMinn, AK Ogawa, Y Wu, J Liu, PG Schultz, FE Romesberg (1999) J Am Chem Soc 121:11585-11586
Ongoing since 1999, the lab has worked to develop infrared probes of protein dynamics, specifically the carbon-deuterium (C-D) bond. We've used C-D bonds to characterize stability, folding, and function of a variety of proteins, and compared their use to other common IR probes of proteins. A second biophysical project focuses on protein evolution, specifically evolution of antibodies and the role of somatic mutations in altering antibody structure and dynamics. We've characterized antibodies raised against several chromophoric antigens, which have allowed us to measure the rigidity of the antibody-antigen complex using nonlinear optical spectroscopy.

We apply spectroscopic methods to characterize the photophysics and dynamics of anti-MPTS antibodies. X-ray crystallography identifies stacking interactions with MPTS that mediate the observed dynamics and provides insight into how dynamics contributes to molecular recognition.
R Adhikary, J Zimmermann, RL Stanfield, IA Wilson, M Oda, FE Romesberg (2019) Biochemistry, published online 19 June
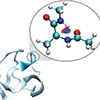
We report a combined atoms in molecules and noncovalent interaction theoretical analysis of electron density that unambiguously supports the characterization of interactions in nSH3 between backbone N atoms and the N–H of the following residue as H-bonds.
M Holcomb, R Adhikary, J Zimmermann, FE Romesberg (2018) J Phys Chem A 122:446–450
ACS Editors' Choice We introduce C-D bonds within different structural elements in the Drosophila transcription factor bicoid, a homeodomain protein with morphogenic activity. C-D absorptions are characterized in the free and DNA-bound states, as well as during thermal denaturation. Residues in the DNA recognition helix, but not other helices, exhibit multiple conformations, and overall the data are consistent with the existence of multiple stable conformations in the folded state.
R Adhikary, YX Tan, J Liu, J Zimmermann, M Holcomb, C Yvellez, PE Dawson, FE Romesberg (2017) Biochemistry 56:2787–2793
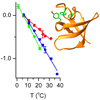
In both model systems and the N-terminal Src homology 3 domain of Crk-II (nSH3), we show that the absorption frequency of cyano and thiocyano probes is linearly correlated with temperature and that the slope of the resulting line (frequency-temperature line slope or FTLS) reflects the nature of the probe’s microenvironment, including whether or not the probe is engaged in H-bonds.
R Adhikary, J Zimmermann, PE Dawson, FE Romesberg (2015) Anal Chem 87:11561–11567
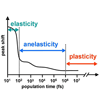
In this work, three affinity matured anti-MPTS antibodies (Abs) are characterized via X-ray and spectroscopic studies and the results are discussed in terms of the elasticity, anelasticity, and plasticity of the complexes. We find that the level of plasticity of the Ab-MPTS complexes is correlated with specificity, suggesting that the optimization of protein dynamics may have contributed to affinity maturation.
R Adhikary, W Yu, M Oda, T Chen, R Walker, R Stanfield, I Wilson, J Zimmermann, FE Romesberg (2015) Biochemistry 54:2085–2093
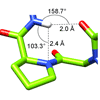
We characterize the IR stretching frequencies of deuterated variants of proline and four proline residues of an Src homology 3 domain protein. CD stretching frequencies are shifted to lower energies due to hyperconjugation with Ni electron density, and along with DFT calculations, the data reveal that the Ni+1-H--Ni interactions constitute H-bonds that may play an important role in protein folding, structure, and function.
R Adhikary, J Zimmermann, J Liu, R Forrest, T Janicki, PE Dawson, S Corcelli, FE Romesberg (2014) J Am Chem Soc 136:13474-13477
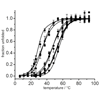
We use IR spectroscopy to characterize nSH3 variants labeled with CN or N3 at five different positions. Like carbon-deuterium (C-D) bonds, CN and N3 can provide information about their surrounding protein environment, but unlike C-D, they tend to be perturbative and thus should be used with caution.
R Adhikary, J Zimmermann, PE Dawson, FE Romesberg (2014) ChemPhysChem 15:849-853
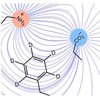
Here, we use the IR absorptions of carbon-deuterium bonds site-selectively incorporated throughout the N-terminal SH3 domain from the murine adapter protein CrkII to characterize their different microenvironments with high spatial and temporal resolution.
R Adhikary, J Zimmermann, J Liu, PE Dawson, FE Romesberg (2013) J Phys Chem B 117:13082-13089
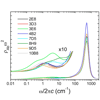
We report the sequence, thermodynamic, and time-resolved spectroscopic characterization of a panel of eight antibodies (Abs) elicited to a chromophoric antigen (MPTS). Three of the Abs arose from unique germline Abs, while the remaining five comprise two sets of siblings that arose by somatic mutation of a common precursor.By characterizing both flexibility and conformational heterogeneity, we show that point mutations are capable of fixing significant differences in protein dynamics.
R Adhikary, W Yu, M Oda, J Zimmermann, FE Romesberg (2012) J Biol Chem 287:27139-27147
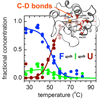
We report a residue-specific characterization of the thermal unfolding mechanism of cytochrome c using C-D bonds site-specifically incorporated at residues dispersed throughout three different structural elements within the protein. Elucidation of the detailed unfolding mechanism and the structure of the associated molten globule, both of which represent challenges to conventional techniques, are highlights of the utility of the C-D technique.
W Yu, P Dawson, J Zimmermann, FE Romesberg (2012) J Phys Chem B 116:6397-6403.
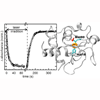
The cyano group is sensitive to its environment, absorbs in a unique region of the protein IR spectrum, and may be appended to an amino acid. Using both steady-state and time-resolved methods, we explore the use of cyano groups as probes of protein microenvironments and dynamics in variants of cytochrome c. We find that the cyano group is a useful site-specific probe of protein microenvironments and dynamics, but that it can also perturb its environment and destabilize the folded state of the protein.
J Zimmermann, MC Thielges, YJ Seo, PE Dawson, FE Romesberg (2011) Angew Chem Int Ed 50:8333-8337
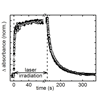
We explore the use of carbon-deuterium (C-D) bonds under time-resolved conditions to follow the unfolding of cytochrome c from a photostationary state that accumulates after CO is photodissociated from the protein’s heme prosthetic group. Our results clearly show that C-D bonds are well-suited to characterize protein folding and dynamics.
J Zimmermann, MC Thielges, W Yu, PE Dawson, FE Romesberg (2011) J Phys Chem Lett 2:412-416
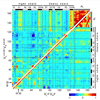
We show that 3PEPS spectroscopy in combination with molecular dynamics simulations can provide a detailed description of protein dynamics for the fluorescein Ab 4−4−20, and how it is evolved for biological function.
J Zimmermann, FE Romesberg, CL Brooks III, IF Thorpe (2010) J Phys Chem B 114:7359-7370
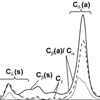
We present the first time-resolved experiments characterizing the population and dephasing dynamics of selectively excited C−D bonds in a deuterated amino acid using both three-pulse photon echo and transient grating experiments. We show that the steady-state C−D absorption linewidths in the deuterated amino acid are broadened by both homogeneous and inhomogeneous effects, and that IVR occurs on a subpicosecond time scale and is nonstatistical.
J Zimmermann, K Gundogdu, ME Cremeens, JN Bandaria, GT Hwang, MC Thielges, CM Cheatum, FE Romesberg (2009) J Phys Chem B 113:7991-7994
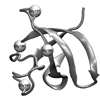
We demonstrate that deuterium atoms incorporated at Cα backbone positions (Cα−D bonds) are sensitive to the local backbone structure. DFT calculations are used to predict that Cα−D bonds of glycine are sensitive to their local structure, with the absorptions red-shifted for an extended β-sheet relative to γ- and α-helix-like turns. These predictions are confirmed in glycine-deuterated variants of the N-terminal Src homology 3 (nSH3) domain protein.
ME Cremeens, J Zimmermann, W Yu, PE Dawson, FE Romesberg (2009) J Am Chem Soc 131:5726-5727
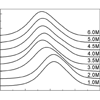
We address the processes that occur upon folding of reduced cyt c induced by photodissociation of CO from the CO-bound, unfolded protein by monitoring the CO vibration with step-scan FT-IR spectroscopy.
MT Thielges, J Zimmermann, FE Romesberg (2009) J Am Chem Soc 131:6054-6055
Using site-selectively incorporated carbon–deuterium bonds, we show that like equine cyt c, bovine cyt c is induced to unfold by guanidine hydrochloride via a stepwise mechanism, but it does not populate an intermediate as is observed with the equine protein.
MC Thielges, J Zimmermann, PE Dawson, FE Romesberg (2009) J Mol Biol 24:159-167
Site-specific deuteration and FT-IR studies reveal that Tyr100 in DHFR plays an important role in catalysis, with a strong electrostatic coupling occuring between Tyr100 and the charge that develops in the hydride-transfer transition state.
D Groff, MC Thielges, S Celliti, PG Schultz, FE Romesberg (2009) Angew Chem Int Ed 48:3478-3481
A grand canonical formalism is developed to combine discrete simulations for chemically distinct species in equilibrium. Each simulation is based on a perturbed funneled landscape. The formalism is illustrated using the alkaline-induced transitions of cyt c as observed by FT-IR spectroscopy and with various other experimental approaches.
P Weinkam, FE Romesberg, PG Wolynes (2009) Biochemistry 48:2394-2402
We demonstrate that the C-D transitions of the deuterated amino acid, leucine-d10 can serve as convenient structural reporters via dual-frequency 2DIR methods, and we discuss the potential of leucine-d10 and other amino acids with deuterium-labeled side chains as probes of protein structure and dynamics.
SRG Naraharisetty, VM Kasyanenko, J Zimmermann, MC Thielges, FE Romesberg, IV Rubtsov (2009) J Phys Chem B 113:4940-4946
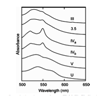
We use acombination of experiments and simulations to provide a high-resolution view of the structural changes in cyt c with increasing alkaline conditions. Alkaline-induced transitions were characterized under equilibrium conditions by monitoring IR absorptions of carbon−deuterium chromophores incorporated at Leu68, Lys72, Lys73, Lys79, and Met80. The data suggest that at least four intermediates are formed as the pH is increased prior to complete unfolding of the protein.
P Weinkam, J Zimmermann, LB Sagle, S Matsuda, PE Dawson, PG Wolynes, FE Romesberg (2008) Biochemistry 47:13470-13480
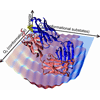
We examine a panel of antibodies elicited to the chromophoric antigen fluorescein. On the basis of isothermal titration calorimetry, we select six antibodies that bind fluorescein with diverse binding entropies, suggestive of varying contributions of dynamics to molecular recognition. We find that more than half of all the somatic mutations among the six antibodies are located in positions unlikely to contact fluorescein directly, and using 3PEPS and TG spectroscopy, we find a high level of dynamic diversity among the antibodies.
MC Thielges, J Zimmermann, W Yu, M Oda, FE Romesberg (2008) Biochemistry 47:7237-7247
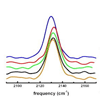
We report a preliminary analysis of (methyl-d3) methionine residues, Met16, Met20, and Met42, within DHFR. The results confirm the sensitivity of the carbon−deuterium bonds to their local protein environment and demonstrate that dihydrofolate reductase is electrostatically and dynamically heterogeneous.
MC Thielges, DA Case, FE Romesberg (2008) J Am Chem Soc 130:6597-6603

We review the role of activation-induced cytidine deaminase (AID) in the complex process of somatic hypermutation (SHM) and present recent advances in experimental methods to characterize antibody dynamics as a function of SHM to help elucidate the role that the AID-induced mutations play in tailoring molecular recognition.
MF Goodman, MD Schaarf, FE Romesberg (2007) Adv Immunol 94:127-155
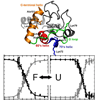
We report the use of a recently developed technique to study the folding of cyt c that is based on the site-selective incorporation of carbon−deuterium (C−D) bonds and their characterization by IR spectroscopy.
LB Sagle, J Zimmermann, PE Dawson, FE Romesberg (2006) J Am Chem Soc 128:14232-14233
We explore the viability of α-carbon deuterated bonds (Cα−D) as infrared (IR) probes of protein backbone dynamics.
CS Kinnaman, ME Cremeens, FE Romesberg (2006) J Am Chem Soc 128:13334-13335
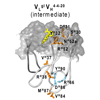
We report the characterization of protein heterogeneity and dynamics as a function of evolution for the antifluorescein antibody 4-4-20. Using nonlinear laser spectroscopy, surface plasmon resonance, and molecular dynamics simulations, we demonstrate that evolution localized the Ab-combining site from a heterogeneous ensemble of conformations to a single conformation by introducing mutations that act cooperatively and over significant distances to rigidify the protein.
J Zimmermann, EL Oakman, IF Thorpe, X Shi, P Abbyad, CL Brooks III, SG Boxer, FE Romesberg (2006) Proc Natl Acad Sci USA 103:13722-13727
We incorporated carbon−deuterium bonds throughout cyt c and characterized their absorption frequencies and line widths using IR spectroscopy. Our data suggest that while the oxidized protein is not more flexible than the reduced protein, it is more locally unfolded.
LB Sagle, J Zimmermann, S Matsuda, PE Dawson, FE Romesberg (2006) J Am Chem Soc 128:7909-7915
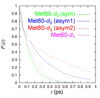
We report the first IR characterization of a single C−D bond within a protein, methyl-d1 Met80 of horse heart cytochrome c. A comparison was made to methyl-d1/d3 methionine as well as methyl-d3 Met80.
ME Cremeens, H Fujisaki, Y Zhang, J Zimmermann, LB Sagle, S Matsuda, PE Dawson, JE Straub, FE Romesberg (2006) J Am Chem Soc 128:6028-6029
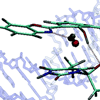
We show that when HBO is positioned in the minor groove of DNA, the enol tautomer of HBO is preferentially stabilized, whereas our previous study showed that when HBO is positioned in the major groove, the keto tautomer is preferentially stabilized. Based on MD simulations, we suggest that this results from the formation of a stable hydrogen-bond between the HBO enol and an H-bond acceptor that is only available in the minor groove.
F-Y Dupradeau, DA Case, C Yu, R Jimenez, FE Romesberg (2005) J Am Chem Soc 127:15612-15617
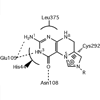
E. coli DNA photolyase is a monomeric light-harvesting enzyme that utilizes a methenyltetrahydrofolate (MTHF) antenna cofactor to harvest light energy for the repair of thymine dimers in DNA. Here, we investigate the origins of how the enzyme tunes the photophysical properties of the antenna cofactor appropriately for biological function.
AA Henry, R Jimenez, D Hanway, FE Romesberg (2004) Chembiochem 5:1088-1094
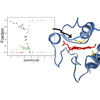
To generate residue-specific information on the equilibrium folding of cyt c, we have semisynthesized the protein with specifically deuterated residues. Plotted as a function of added guanidine hydrochloride denaturant, the absorption intensities of the protein C-D bonds reveal that cyt c undergoes a conformational change at the protein-based heme ligand, Met80, which is then followed by a more global unfolding at 2.3 M GdnHCL.
LB Sagle, J Zimmermann, PE Dawson, FE Romesberg (2004) J Am Chem Soc 126:3384-3385
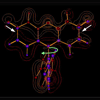
3PEPS and steady-state spectroscopy in combination with binding and structural data, are used to examine the immunological evolution of protein flexibility in an anti-fluorescein antibody. We show that antibody dynamics are systematically manipulated during affinity maturation, and suggest that the evolution of protein flexibility may be a central component of the immune response.
R Jimenez, G Salazar, J Yin, T Joo, FE Romesberg (2004) Proc Natl Acad Sci USA 101:3803-3808

We use photon echo spectroscopy to measure the response of three antibody-binding sites to perturbation from electronic excitation of a bound antigen, fluorescein. The three antibodies show motions that range in time scale from tens of femtoseconds to nanoseconds. View the cover story.
R Jimenez, G Salazar, KK Baldridge, FE Romesberg (2003) Proc Natl Acad Sci USA 100:92-97

We find that the typical time for the excited-state intramolecular proton-transfer reaction of the syn-enol tautomer in solution and in DNA is 150 fs. Picosecond rise and decay components in the fluorescence transients with characteristic times between 3 and 25 ps are observed and attributed to the effects of vibrational cooling.
H Wang, H Zhang, OK Abou-Zied, C Yu, FE Romesberg, M Glasbeek (2003) Chem Phys Lett 367:599-608
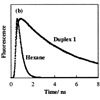
Transient absorption, transient grating, and 3PEPS measurements are used to characterize the photophysics of the heme chromophore in the folded protein and in two different unfolded proteins. We find that cyt c has little structural heterogeneity in the folded state, and a larger degree of structural heterogeneity in the unfolded states that depends on the unfolding conditions.
R Jimenez, FE Romesberg (2002) J Phys Chem B 106:9172-9180
The solvent-dependent ground-state conformational equilibrium and excited-state dynamics of 2-(2‘-hydroxyphenyl)benzoxazole are characterized in several solvents on the femtosecond to nanosecond time scales.
OK Abou-Zied, R Jimenez, EHZ Thompson, DP Millar, FE Romesberg (2002) J Phys Chem A 106:3665-3672
The dynamics of the protein-based methionine heme ligand were examined by selectively deuterating the ligand's methyl group. The frequency and line width of the C−D bonds were easily observable and shown to be sensitive to mutation-induced changes in the protein redox potential.
JK Chin, R Jimenez, FE Romesberg (2002) J Am Chem Soc 124:1846-1847

The reorganization energies of the chromophore−antibody complexes are determined and are found to be distributed over a wide range. 3PEPS spectroscopy is used to measure the time scales of the reorganization process in one complex. We find that the antibody motions occur on time scales ranging from 75 femtoseconds to 67 picoseconds.
R Jimenez, DA Case, FE Romesberg (2002) J Phys Chem B 106:1090-1103
HBO was synthesized and incorporated into DNA oligonucleotides and each oligonucleotide was hybridized to a complementary oligonucleotide containing an abasic site at the position opposite HBO. Analysis of steady state emission spectra of the oligonucleotides indicates that the DNA environment selectively stabilizes the proton-transfer product tautomer of HBO, suggesting that flanking bases may be strongly coupled electronically.
OK Abou-Zied, R Jimenez, FE Romesberg (2001) J Am Chem Soc 106:1090-1103
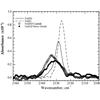
We introduce C−D bonds into horse heart cyt c and use the C−D stretching vibration, which absorbs at approximately 2100 cm-1, as a spectroscopic probe of the protein environment.
JK Chin, R Jimenez, FE Romesberg (2001) J Am Chem Soc 123:2426-2427
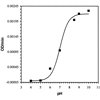
Hammett correlation studies of the aldol and retroaldol reactions catalyzed by antibodies 38C2 and 24H6 reveal that although the two antibodies have broad substrate specificities, they utilize slightly different mechanisms. Antibody 38C2 adopts a mechanism that is reminiscent of an acid-catalyzed aldol reaction, while antibody 24H6 follows a mechanism that is similar to the base-catalyzed reaction.
H Shulman, C Makarov, AK Ogawa, FE Romesberg, E Keinan (2000) J Am Chem Soc 122:10743-10753
2-(2‘-Hydroxyphenyl)benzoxazole (HBO) undergoes rapid photoinduced proton transfer from the enol-imine to the keto-amine tautomer. When incorporated in duplex DNA opposite an abasic site, HBO appears to be a good mimic of a natural DNA base pair based on duplex stability, UV and CD spectroscopy, and molecular dynamics simulations.
AK Ogawa, OK Abou-Zied, V Tsui, R Jimenez, DA Case, FE Romesberg (2000) J Am Chem Soc 122:9917–9920
Back in the early 2000s, our work to combat bacteria was focused on understanding DNA damage repair and the inhibition of the bacterial SOS response. We also worked for a time on the equivalent biochemical pathways in yeast. More recently, work has focused on the arylomycin family of natural products, which targets the bacterial enzyme signal peptidase, and the biochemical consequences of signal peptidase inhibition.

We identify the proteins whose extracytoplasmic localization is inhibited by arylomycin in E. coli. We find among these proteins a number of virulence factors and show that even at sub-MIC levels, the arylomycins inhibit the formation of flagella, and reduce motility, biofilm formation, and the dissemination of antibiotic resistance through horizontal gene transfer.
SI Walsh, DS Peters, PA Smith, A Craney, MM Dix, BF Cravatt, FE Romesberg. (2019) Antimicrob Agents Chemother 63:e01253-18

We report the synthesis and characterization of the first 5S β-lactams to possess significant antibacterial activity and the first β-lactams imparted with antibacterial activity via the optimization of inhibition of a target other than a PBP.
CH Yeh, SI Walsh, A Craney, MG Tabor, AF Voica, R Adhikary, SE Morris, FE Romesberg (2018) ACS Med Chem Lett 9:376–380
A new route to a formal synthesis of the arylomycins and a collection of new analogues along with their biological evaluation is reported.
DS Peters, FE Romesberg, PS Baran (2018) J Am Chem Soc 140:2072–2075

We further characterize the S. aureus operon ayrRABC and show that the inducing signal for ayrRABC depression is accumulation of a subset of preproteins with signal peptides that are stable toward further processing. The results elucidate the mechanism by which S. aureus monitors and responds to secretion stress.
A Craney, FE Romesberg (2017) mBio, 8:e01507-17
We identify the S. aureus operon ayrRABC and show that once released from repression by AyrR, the protein products AyrABC together confer resistance to the SPase inhibitor arylomycin M131 by providing an alternate and novel method of releasing translocated proteins, and we demonstrate that AyrABC functionally complements SPase by mediating the processing of the normally secreted proteins. Overall, the data demonstrate that ayrRABC encodes a secretion stress-inducible alternate terminal step of the general secretory pathway.
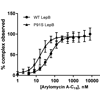
A Craney, MM Dix, R Adhikary, BF Cravatt, FE Romesberg (2015) mBio 6:e01178-15
We demonstrate that arylomycin activity is conserved against a broad range of Y. pestis strains and that this activity results from SPase inhibition. The origins of the sensitivity are traced to an increased dependence on SPase that results from high levels of protein secretion under physiological conditions.
DB Steed, J Liu, E Wasbrough, L Miller, S Halasohoris, J Miller, B Somerville, JR Hershfield, FE Romesberg (2015) Antimicrob Agents Chemother 59:3887-3898
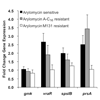
We characterized the susceptibility of a panel of S. aureus strains to two arylomycin derivatives and observed a wide range of susceptibilities. We found that resistant strains were sensitized by co-treatment with tunicamycin, which inhibits the first step of wall teichoic acid synthesis. Transcriptional profiling using growth inhibitory concentrations of arylomycin revealed further insight into how this pathogen copes with secretion stress.
A Craney, FE Romesberg (2015) Antimicrob Agents Chemother 59:3066-3074
In this collaborative work with the Baran Lab, we show that the axinellamines have promising activity against Gram-positive and Gram-negative bacteria, suggesting that their scaffold has the potential for further development. Details regarding their mode of action remain to be elucidated, but the axinellamines appear to cause secondary membrane destabilization and may inhibit normal septum formation.
RA Rodriguez, DB Steed, Y Kawamata, S Su, PA Smith, TC Steed, FE Romesberg, PS Baran (2014) J Am Chem Soc 136:15403-15413
We report the synthesis and evaluation of several arylomycin derivatives, and demonstrate that both C-terminal homologation with a glycyl aldehyde and addition of a positive charge to the macrocycle increase the activity and spectrum of the arylomycin scaffold.
J Liu, PA Smith, DB Steed, FE Romesberg (2013) Bioorg Med Chem Lett 23:5654-5659

We examine arylomycin activity as a function of concentration, bacterial cell density, target expression levels, and bacterial growth phase. We find that the activity of the arylomycins results from an insufficient flux of proteins through the secretion pathway and the resulting mislocalization of proteins. We also demonstrate that SPase inhibition results in synergistic sensitivity when combined with an aminoglycoside.
PA Smith, FE Romesberg (2012) Antimicrob Agents Chemother 56:5054-5060

Stressful lifestyle associated mutation (SLAM) assays are used to demonstrate that both S. cerevisiae and C. albicans undergo mutations to acquire resistance to the antifungal agents 5-fluorocytosine and caspofungin. Examination of the mutation spectrum shows that these aquired mutations are different from those that the occur spontaneously, in the absence of antifungal treatment. Overall, the work present a much needed model system for studying adaptive mutagenesis in eukaryotes.
D Quinto-Alemany, A Canerina-Amaro, LG Hernández-Abad, F. Machín, FE Romesberg, C Gil-Lamaignere (2012) PLoS ONE 7:e42279

We use an arylomycin as a probe of protein secretion to identify forty-six proteins whose extracellular accumulation in S. aureus requires the activity of signal peptidase (SPase). These include a wide variety of virulence proteins with identifiable Sec-type signal peptides, as well as some proteins with non-canonical cleavage sites.
MA Schallenberger, S Niessen, C Shao, BJ Fowler, FE Romesberg (2012) J Bacteriol 194:2677-2686
We report the first total synthesis of a lipoglycopeptide member of the arylomycin family of natural products, and we demonstrate that it effectively penetrates Gram-negative bacteria and is limited by the same resistance mechanism as other members of the arylomycin class of antibiotics (i.e. mutation in the target protein signal peptidase (SPase)). Unlike the other arylomycins, the lipoglycopeptides are glycosylated, and the structural data reveal that this sugar is most solvent exposed when in complex with SPase.
J Liu, C Luo, PA Smith, JK Chin, MGP Page, M Paetzel, FE Romesberg (2011) J Am Chem Soc 133:17869-17877
A small library of 2′,4′- and 3′,4′-bridged nucleoside analogues was synthesized and tested for antibacterial, antitumor, and antiviral activities, leading to the identification of one nucleoside that exhibited significant antiviral activity against pseudoviruses SF162 (IC50 = 7.0 μM) and HxB2 (IC50 = 2.4 μM). These findings render bridged nucleosides as credible leads for drug discovery in the anti-HIV area of research.
KC Nicolaou, SP Ellery, F Rivas, K Saye, E Rogers, TJ Workinger, M Schallenberger, R Tawatao, A Montero, A Hessel, F Romesberg, D Carson, D Burton (2011) Bioorg Med Chem 19:5648-5669
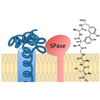
Resistance to the arylomycins in several key human pathogens is due to the presence of a specific Pro residue in the target peptidase that disrupts interactions with the lipopeptide tail of the antibiotic. Here, we describe the synthesis and characterization of arylomycin derivatives with altered lipopeptide tails, including several with an increased spectrum of activity against S. aureus.
TC Roberts, M. Schallenberger, J Liu, PA Smith, FE Romesberg (2011) J Med Chem 54:4954-4963
We report the total synthesis of arylomycin B-C16, and its aromatic amine derivative. While the aromatic amine loses activity against all bacteria tested, the B series compound shows activities that are similar to the A series compounds, except that it also gains activity against the important pathogen Streptococcus agalactiae.
TC Roberts, PA Smith, FE Romesberg (2011) J Nat Prod 74:956-961
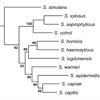
We report the activity of a synthetic arylomycin derivative against clinical isolates of coagulase-negative staphylococci (CoNS). Against many important CoNS, e.g. S. epidermidis, S. haemolyticus, S. lugdunensis, and S. hominis, we find that the arylomycin has activity equal to or greater than that of vancomycin, the antibiotic most commonly used to treat CoNS infections.
PA Smith, ME Powers, TC Roberts, FE Romesberg (2011) Antimicrob Agents Chemother 55:1130-1134

This work describes the use of an arylomycin derivative, along with two-dimensional gel electrophoresis and LC-MS/MS, to identify eleven proteins whose secretion from stationary phase S. epidermidis is dependent on SPase activity. The data also suggest that the arylomycins should be valuable chemical biology tools for the study of protein secretion in a wide variety of different bacteria.
ME Powers, PA Smith, TC Roberts, BJ Fowler, CC King, SA Trauger, G Siuzdak, FE Romesberg (2011) J Bacteriol. 193:340-348
This work demonstrates that the apparently narrow spectrum of the arylomycin natural products results not from intrinsic limitations, but from target mutations and that the arylomycins have a much broader spectrum of activity than previously thought.
PA Smith, TC Roberts, FE Romesberg (2010) Chem Biol 17:1223-1231
Plants provide a unique source of diverse secondary metabolites with potentially important biological activities, with one of the most promising classes being the indole alkaloids. One such indole alkaloid natural product is psychotrimine, which has attracted considerable interest from the synthetic and medicinal chemistry communities owing to its unusual connectivity between tryptamine subunits. Here, we examine the potential antibacterial activity of this unique indole alkaloid.
MA Schallenberger, T Newhouse, PS Baran, FE Romesberg (2010) J Antibiot 63:685-687

CPAF is secreted into host cytosol to degrade various host factors for benefiting chlamydial intracellular survival. Here, we show experimental evidence that the chlamydial virulence factor CPAF relies on sec-dependent transport for crossing the chlamydial organism inner membrane.
D Chen, L Lei, C Lu, R Flores, MP DeLisa, TC Roberts, FE Romesberg, G Zhong (2010) Microbiology 156:3031-3040

Epithelial cells are highly regarded as the first line of defense against microorganisms, but the mechanisms used to control bacterial diseases are poorly understood. A component of the DNA damage repair regulon, SulA, is essential for UPEC virulence in a mouse model for human urinary tract infection, suggesting that DNA damage is a key mediator in the primary control of pathogens within the epithelium. In this study, we examine the role of DNA damage repair regulators in the intracellular lifestyle of UPEC within superficial bladder epithelial cells.
B Li, P Smith, DJ Horvath Jr, FE Romesberg, SS Justice (2010) Microb Infect 12:662-668
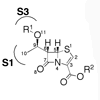
We report the first synthesis of a 5S penem, known to bind bacterial type I signal peptidase, from the commercially available and inexpensive 6-aminopenicillanic acid, and we demonstrate the first in vivo activity of the compound and use structure–activity relationship studies to begin to define the determinants of signal peptidase binding and to begin to optimize the penem as an antibiotic.
DA Harris, ME Powers, FE Romesberg (2009) Bioorg Med Chem Lett 19:3787-3790

To further understand the mutagenic response to DNA damage, we screen a collection of 4848 haploid gene deletion strains of S. cerevisiae for decreased damage-induced mutation of the CAN1 gene, and we identify a pathway of induced mutation that involves a replicative polymerase and that is effectively inhibited by the RNR inhibitor, hydroxyurea.
ET Lis, BM O'Neill, C Gil-Lemaignere, JK Chin, FE Romesberg (2008) DNA Repair 7:801-810
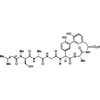
We report the synthesis and characterization of an arylomycin, a natural product inhibitor of bacterial signal peptidase.
TC Roberts, PA Smith, RT Cirz, FE Romesberg (2007) J Am Chem Soc 129:15830-15838
We present genetic and biochemical evidence that the type 2A-like protein phosphatase Pph3 forms a complex with Psy2 (Pph3–Psy2) that binds and dephosphorylates activated Rad53 during treatment with, and recovery from, methylmethane sulfonate-mediated DNA damage. Ourfindings suggest that Rad53 regulates replication fork restart and initiation of late firing origins independently and that regulation of these processes is mediated by specific Rad53 phosphatases.
BM O'Neill, SJ Szyjka, ET Lis, AO Bailey, JR Yates III, OM Aparicio, FE Romesberg (2007) Proc Natl Acad Sci USA 104:9290-9295

We characterize the global transcriptional response of S. aureus to ciprofloxacin and find that the drug induces prophage mobilization as well as significant alterations in metabolism, most notably the up-regulation of the tricarboxylic acid cycle.[Download Supporting Information]
RT Cirz, MB Jones, TD Minogue, B Jarrahi, SN Peterson, FE Romesberg (2007) J Bacteriol 189:531-539

We use DNA microarrays to characterize the global transcriptional response of P. aeruginosa to clinical-like doses of the antibiotic ciprofloxacin and also to determine the component that is regulated by LexA cleavage and the SOS response.[Download Supporting Data][Download SI Methods & Tables]
RT Cirz, BM O'Neill, JA Hammond, SR Head, FE Romesberg (2006) J Bacteriol 188:7101-7110

Here, we describe S. cerevisiae Doa1, which helps to control the damage response by channeling ubiquitin from the proteosomal degradation pathway into pathways that mediate altered DNA replication and chromatin modification. Our data suggest that Doa1 is the major source of ubiquitin for the DNA damage response and that Doa1 also plays an additional essential and more specific role in the monoubiquitination of histone H2B.
ET Lis, FE Romesberg (2006) Mol Cell Biol 26:4122-4133

We show that Esc4 interacts with several proteins involved in the repair and processing of stalled or collapsed replication forks, including the recombination protein Rad55, and we propose that Esc4 associates with ssDNA of stalled forks and acts as a scaffolding protein to recruit and/or modulate the function of other proteins required to reinitiate DNA synthesis.
JK Chin, VI Bashkirov, W-D Heyer, FE Romesberg (2006) DNA Repair 5:618-628

We show that LexA cleavage and polymerase derepression are required for the evolution of clinically significant resistance in MMR-defective E. coli.
RT Cirz, FE Romesberg (2006) Antimicrob Agents Chemother 50:220-225

Read about this article in C&EN, Nature Reviews Genetics, or listen to a Scientific American podcast
RT Cirz, JK Chin, DR Andes, V de Crécy-Lagard, WA Craig, FE Romesberg (2005) PLoS Biol 3:e176
The stabilization and processing of stalled replication forks is required to maintain genome integrity in all organisms. Based on both physical and genetic interactions detected between WSS1, PSY2, and TOF1, we suggest that Wss1 and Psy2 similarly function to stabilize or process stalled or collapsed replication forks.
BM O'Neill, D Hanway, EA Winzeler, FE Romesberg (2004) Nucleic Acids Res 32:6519-6530

A competitive growth assay is used to identify yeast genes involved in the repair of UV- or MMS-induced DNA damage using a collection of 2,827 yeast strains in which each strain has a single ORF replaced with a cassette containing two unique sequence tags. Identified genes include three uncharacterized ORFs, as well as genes that encode protein products implicated in ubiquitination, gene silencing, and transport across the mitochondrial membrane.
D Hanway, JK Chin, G Xia, G Oshiro, EA Winzeler, FE Romesberg (2002) Proc Natl Acad Sci USA 99:10605-10610
Not interested in every detail? Read a review instead.

We review recent work from the Benner lab with DNA that is made up of the natural letters dG, dC, dA, and dT, as well as their unnatural letters, dP, dZ, dS, and dB.
VT Dien, M Holcomb, FE Romesberg (2019) Biochemistry 58:2581-2583

We review progress and achievements made towards developing a functional unnatural base pair and its use to generate semisynthetic organisms.
VT Dien, SE Morris, RJ Karadeema, FE Romesberg (2018) Curr Opin Chem Biol 46:196-202
We review the work that led to the creation of semi-synthetic organisms able to produce proteins via the decoding of a six-letter genetic alphabet.
Y Zhang, FE Romesberg (2018) Biochemistry, 57:2177–2178
ACS Editors' Choice We review our efforts to develop unnatural base pairs (UBPs) that ultimately culminated in the discovery of dNaM-dTPT3, as well as a family of related UBPs, all of which have been used to create semi-synthetic organisms.
A Feldman, FE Romesberg (2018) Acc Chem Res 51:394–403
We review our recent work describing how without any sequence-specific optimization, PCT is capable of producing 103–105 copies of RNA from a single strand of DNA, far in excess of what is possible with conventional transcription. We also discuss how PCT is especially efficient with shorter RNAs, as well as with 2′-F-modified triphosphates.
T Chen, FE Romesberg (2017) Biochemistry 56:5227–5228
We review the strengths and weaknesses of the different transparent window vibrational probes, methods by which they may be site-specifically incorporated into peptides and proteins, and the physicochemical properties they may be used to study. These topics are put into context through four case studies focused on KSI, SH3, DHFR, and cyt c.
R Adhikary, J Zimmermann, FE Romesberg (2017) Chem Rev 117:1927–1969

We serve as guest editors for a special issue of Bioorg. Med. Chem. In this preface, we give a bit of background on drug discovery and the impending crisis posed by antibiotic resistance, as well as an overview of the contributed articles.
A Craney, FE Romesberg (2016) Bioorg Med Chem 24:6225-6226
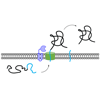
We review how the inhibition of SPase may affect bacterial virulence, and how SPase itself contributes to functions beyond mediating bacterial secretion.
SI Walsh, A Craney, FE Romesberg (2016) Bioorg Med Chem 24:6370-6378
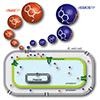
We review nucleotide modifications, such as those to phosphate and sugar moieties that increase nuclease resistance or the range of activities possible, as well as whole nucleobase replacement that results in selective pairing and the creation of unnatural base pairs. Both in vitro and in vivo examples are discussed, including efforts to create semi-synthetic organisms with altered or expanded genetic alphabets.
T Chen, N Hongdilokkul, Z Liu, D. Thirunavukarasu, FE Romesberg (2016) Curr Opin Chem Biol 34:80–87
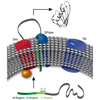
We review the information gained from the use of SPase inhibitors as probes of prokaryote biology. A thorough understanding of the consequences of SPase inhibition and the mechanisms of resistance that arise are essential to the success of SPase as an antibiotic target. In addition to the role of SPase in processing secreted proteins, the use of SPase inhibitors has elucidated a previously unknown function for SPase in regulating cleavage events of membrane proteins.
A Craney, FE Romesberg (2015) Biooog Med Chem Lett 25:4761–4766

Natural nucleic acids and the genetic information they encode are limited by the use of only four nucleotides that form two base pairs, (d)G-(d)C and d(A)-dT/U. In the past decade, three classes of unnatural base pairs have been developed to a high level of proof-of-concept. This review summarizes their development and the potentially revolutionary applications that they are now enabling.
DA Malyshev, FE Romesberg (2015) Angew Chem Int Ed 54:11930–11944
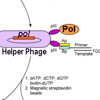
We review directed evolution methods used to tailor the properties and/or substrate repertoire of polymerases for different applications and provide a survey of the many polymerases that have been evolved.
T Chen, FE Romesberg (2014) FEBS Lett 588:219-229

We describe the experimental procedures required to use C-D bonds and FT IR spectroscopy to characterize protein dynamics, structural and electrostatic heterogeneity, ligand binding, and folding.
J Zimmermann, FE Romesberg (2014) Methods Mol Biol 1084:101-119

We provide a concise review of type I signal peptidases, including historical and structural biology information, as well as preparation methods and work to identify inhibitors.
DA Harris, FE Romesberg, Handbook of Proteolytic Enzymes (Third Edition)., ND Rawlings, G Salvesen, Eds. (2013) Volume 3, 3501-3508.

We review the arylomycin family of natural product antibiotics, highlighting how their characterization has provided new insight into how antibiotics evolve in nature.
YX Tan, FE Romesberg (2012) MedChemComm 3:916-925
At low pH, the experimentally observed folding sequence of cytochrome c deviates from that at pH 7 and from models with perfectly funneled energy landscapes. We account for these alternative folding pathways using complex models that begin from native structure-based terms and also add the chemical frustration that arises because some regions of the protein are destabilized more than others due to the heterogeneous distribution of titratable residues that are protonated at low pH.
P Weinkam, J Zimmermann, FE Romesberg, P Wolynes (2010) Acc Chem Res 43:652-660

We review novel experimental methods to characterize conformational heterogeneity and dynamics in proteins.
J Zimmermann, MC Thielges, W Yu, FE Romesberg (2009) in Protein Engineering Handbook, Vol. 1, S. Lutz and U.T. Bornscheuer, Eds., Wiley-VCH:Weinheim, 147-186

We review screening and selection methods to identify polymerases with novel function.
AM Leconte, FE Romesberg, (2009) in Protein Engineering, C. Köhrer and U.L. RajBhandary, Eds., Springer-Verlag:Berlin, 291-314

Editorial comment for the special issue of Current Opinion in Chemical Biology describing several novel approaches to developing therapeutics that target both bacterial and human proteins.
A Mapp, FE Romesberg (2008) Curr Opin Chem Biol 12:387-388
We review progress toward understanding the functions of phosphatases in checkpoint deactivation in S. cerevisiae, focusing on the non-redundant roles of the type 2A phosphatase Pph3 and the PP2C phosphatases Ptc2 and Ptc3 in the deactivation of Rad53.
J Heideker, ET Lis, FE Romesberg (2007) Cell Cycle 6:3058-3064
We review what is known about induced mutagenesis in bacteria as well as evidence that it contributes to the evolution of antibiotic resistance and we discuss the possibility that components of induced mutation pathways might be targeted for inhibition as a novel therapeutic strategy to prevent the evolution of antibiotic resistance.
RT Cirz, FE Romesberg (2007) Crit Rev Biochem Mol Biol 42:341-354
An improved understanding of bacterial stress responses and evolution suggests that in the ability of bacteria to survive antibiotic therapy either by transiently tolerating antibiotics or by evolving resistance may require specific biochemical processes. We review early efforts toward inhibiting these processes as a means to prolong the efficacy of current antibiotics and provide an alternative to escalating the current arms race between antibiotics and bacterial resistance.
PA Smith, FE Romesberg (2007) Nat Chem Biol 3:549-556

Editorial comment on the special biopolymers issue of Curr. Opin. Chem. Biol.
H Bayley, FE Romesberg (2006) Curr Opin Chem Biol 10:598-600
We review the report of the structure of xDNA from the Kool lab.
AM Leconte, FE Romesberg (2006) Nature 444:553-555
We review a report by Hirao et al. demonstrating that DNA containing an unnatural base pair is both efficiently PCR amplified and transcribed.
AM Leconte, FE Romesberg (2006) Nat Meth 3:667-668

We review a report by Prudhomme et al. in Science that demonstrates how Streptococcus pneumoniae may increase their chances of acquiring exogenous DNA through the upregulation of competence in response to antibiotic treatment.
RT Cirz, N Gingles, FE Romesberg (2006) Nat Meth 12:890-891
We review several activity-based screening and selection approaches that have been used to identify polymerases with novel activities.
AA Henry, FE Romesberg (2005) Curr Opin Biotech 16:370-377

We review screening and selection approaches, both in vivo and in vitro, that have been used to evolve polymerases with a variety of novel properties, such as thermal stability, resistance to inhibitors, and altered substrate specificity.
RC Holmberg, AA Henry, FE Romesberg (2005) Biomol Eng 22:39-49
We review the role of quantum tunneling as a mechanism of the enymatic transfer of hydrogenic entities (protons, hydrogen atoms, and hydride ions), including the background of the subject, the conceptual apparatus that underlies the isotopic studies, the phenomenology of the experimental observations, and a qualitative sketch of the interpretative, mechanistic models that have emerged.
FE Romesberg, RL Schowen (2004) Phys Org Chem 39:27-77
This review provides a critical comparison of the third base pair candidates and discusses the further work required to expand the genetic alphabet.
AA Henry, FE Romesberg (2003) Curr Opin Chem Biol 7:727-733
We describe methods for the synthesis and characterization of modified nucleobases.
FE Romesberg, C Yu, S Matsuda, AA Henry (2002) Curr Prot Nucleic Acid Chem, Unit 1.5