

Scientists Solve Protein Structure Revealing Secrets of Cell Membranes
By Laura Bonetta
A team of scientists at The Scripps Research Institute and the National Institutes of Health (NIH) has discovered the structure of a protein that pinches off tiny pouches from cells' outer membranes. Cells use these pouches, or vesicles, to carry nutrients and other essential substances, but many medicines also hitch a ride inside them.
The structure of the protein, called dynamin, is helping to answer many longstanding questions about how vesicles form, advancing knowledge of a process critical to cell survival. The findings may also have implications for designing better ways for delivering drugs.
The research was published on April 28, 2010, in an advance, online issue of the prestigious journal Nature.
The Puzzle of Pinching Off Membranes
The cell membrane typically acts as a barrier around the cell, keeping out harmful materials. But cells also need some substances to get inside.
To get past the membrane, nutrients or hormones in the bloodstream, for example, bind to specific receptors on cells membranes. The membrane then forms a pit around these bound molecules, which is squeezed into a pouch, or vesicle, that detaches from the rest of the cell membrane and carries its essential cargo into the cell. Nerve cells use this same vesicle-making mechanism, called endocytosis, to maintain signaling from one cell to another.
"Endocytosis is how cells communicate," says Sandra Schmid, chair of the Scripps Department of Cell Biology and senior author of the Nature article along with Fred Dyda at NIH. "It's critical for many biological functions from controlling blood pressure to getting rid of glucose."
Despite the importance of endocytosis, scientists have been puzzled by how cells perform this process. But they knew that at least one molecule, dynamin, played a starring role.
Dynamin belongs to a large family of enzymes called GTPases. These enzymes bind a chemical called GTP and convert it to a simpler form (GDP), releasing energy in the process. During this conversion a GTPase undergoes a change in shape, enabling it to perform a particular function—such as making vesicles.
Initially, most scientists believed that many dynamin molecules assembled long spirals on cell membranes, and that in the presence of GTP these spirals tightened, lopping off a vesicle.
But a year ago, a study published in Cell by Schmid's group challenged that view. By watching vesicle transformation through a microscope, the scientists showed that dynamin proteins only form a short collar around the cell membrane. What's more, dynamin can act alone, without the help of any other proteins.
"Dynamin is the master regulator of endocytosis," says Schmid. "It is involved at every stage of vesicle formation."
Seeing Is Believing
That study did not reveal how the dynamin collars pinch off membrane vesicles, though. Thus, Schmid and others turned their attention to dynamin's GTPase activity for clues of how it controls the process.
One way to figure out how a protein functions is to determine its structure. To this end, scientists often use a technique called X-ray crystallography, which involves making crystals of the protein of interest and then bombarding them with X-rays to see the positions of the atoms.
But dynamin is a large molecule, containing almost 1,000 amino acids, making it difficult to crystallize. To overcome the problem, about three years ago Joshua Chappie, then a graduate student in Schmid's laboratory, engineered a shorter version of dynamin that retained the same GTPase activity as the complete protein.
With this shortened protein Chappi quickly obtained crystals and then examined them by X-ray crystallography. However, the resulting data proved impossible to interpret because of a kind of double vision in the X-ray signals. Then came a breakthrough: Chappie discovered that the minimal dynamin formed dimers during its normal cycle of GTP hydrolysis.
Researchers knew that dynamin is found in cells as a group of four molecules, or a tetramer. Like a handful of Tootsie Pops™, dynamin tetramers are held together by long stalk regions with the GTPase domains protruding from their tops. However, the minimal dynamin lacked the stalk regions and exists as monomer. When dynamin assembles into short collars, the GTPase domains of neighboring tetramers form functional dimers that are necessary for GTPase activity and for membrane pinching.
Based on the structure that Chappie, Schmid, Dyda and colleagues described, the scientists suggest that when the GTPase domains from different tetramers pair up the structures of the tetramers shift, making them less stable. The conversion of GTP to GDP then causes another change in shape in the tetramers, possibly through a twisting motion on the membrane. As a result, the dimers dissociate and the entire collar structure comes apart. Vesicle formation probably involves repeated cycles of collar assembly, GTP binding, GTPase domain dimerization, conversion of GTP to GDP, and disassembly of the dynamin collar. These cycles eventually twist and pinch off the membrane.
The crystal structure of the shortened dynamin has revealed other important information. For example, the protein contains three amino acids that are absolutely critical for its GTPase activity and that are conserved among all GTPases with similar functions, providing a "signature" for this group of enzymes.
"Many of the questions we've been trying to answer for the past decade were answered by this structure," says Schmid.
New questions, of course, now follow and the Schmid team is investigating. In addition to Chappie, Schmid, and Dyda, authors of the paper, "G domain dimerization controls dynamin's assembly stimulated GTPase activity," are Sharmistha Acharya and Marilyn Leonard of Scripps Research. See http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature09032.html
The work was supported by the NIH. X-ray diffraction data were obtained at the Advanced Photon Source, Argonne National Laboratory, which is supported by the National Institutes of Health and the U.S. Department of Energy.
Send comments to: mikaono[at]scripps.edu
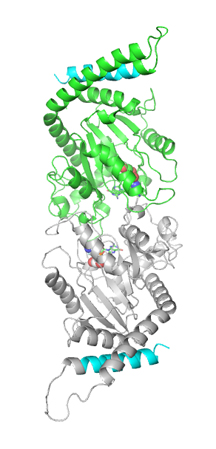
"Many of the questions we've been trying to answer for the past decade were answered by this structure," says Sandra Schmid, chair of the Scripps Department of Cell Biology.
