

Molecular Graphics and Music of the Spheres
By Eric Sauter
Spheres, aside from being a good thing to have at the beach, are nature's carry all. Not only is the sphere the simplest finite surface that encapsulates space, but spheres also have it all over other forms of encapsulation (ignore the cube, eschew the cylinder); they have the smallest enclosed surface area that holds a specific volume and they can hold the biggest volume of all defined enclosed surface areas.
The optimal-use sphere comes in all sizes and appears everywhere—in water drops, air bubbles, and spherical viruses, whose shape is actually a 20-sided (all equilateral triangles) icosahedron.
Now, scientists from The Scripps Research Institute's Molecular Graphics Laboratory have learned how to mimic nature's viral design. They have developed a novel strategy that adheres to the geometric and energetic characteristics of the protective viral icosahedron capsid but that uses synthetic compounds instead of protein subunits.
The work, led by Scripps Research scientists Arthur Olson and Ehud Keinan, was published in the December 26, 2007 (Volume 104, Number 52) edition of the Proceedings of the National Academy of Sciences (PNAS).
The scientists' computer-generated design is based on the self-assembly of synthetic pentagonal tiles. These nanocontainers, which are designed to assemble and disassemble under controlled conditions, have exterior diameters of 25 to 50 Å (angstroms) and interior volumes in the range of 15,000 to 1,000,000 Å3. An angstrom is equal to one ten-billionth of a meter.
The Scripps Research scientists used corannulene, a bowl-shaped molecule that has the ability to turn itself inside out 200,000 times a second. Corannulene is a synthetic compound made up of carbon and hydrogen and consists of a centrally joined circle of five benzene rings; its saucer or concave shape is determined by its chemical bonds. The molecule, which is similar to buckminsterfullerene (used to make the happy-go-lucky-sounding buckyballs; corannulene is also known as the buckybowl because of its saucer shape), can now be used to produce self-assembling capsid-like structures as well, thanks to Scripps Research scientists.
"This is a fairly fundamental study," said Olson, "so it's difficult to say how these structures might ultimately be used. From our point of view, the important finding is that we have shown the ability to construct very specific capsules in this scale range—between buckyballs and viral capsids. We can specify the size and volume, and design them so that they will bind together under certain circumstances and fall apart under others."
The potential applications for these corannulene-based structures range from aiding laboratory studies to clinical therapeutics.
"Because you can encapsulate small amounts of molecular material, these structures could serve as a vehicle for drug delivery," Olson said. "The interesting thing is you can design attachment sites on the outside of the synthetic spheres for specific biological targets—so they become a general-purpose nano-delivery system. The other unique thing about the technology is the fact that the 12 self-assembling tiles don't have to be identical. That gives the potential of having different targets designed into a particular container—like having a dozen different postal codes. This also opens up the possibility of constructing larger and more complex structures."
As an interesting side note, Olson has a history with corannulene; as an undergrad at the University of Michigan in the 1960s, he worked in the laboratory that initially solved the compound's structure.
Viral Self-Assembly
Olson and his colleagues arrived at a general synthetic self-assembly idea by constructing and examining physical models of spherical virus assembly.
"Scientists rarely write about how an idea is generated," he said. "It's one of the most important parts of a study, but the least written about."
In this case, the scientists began by shaking plastic models of the poliovirus components in a container.
Olson built a model of the poliovirus assembly units, a five-sided assembly with electrostatic charges along the five edges, and substituted magnets for the electrostatic charges.
"The idea of shaking them to reproduce self-assembly—I didn't think they would self-assemble, I just wanted them to stick together," he said. "I put a dozen of these plastic tiles in a bottle and shook it. It took about one to two minutes for them to assemble."
Olson then expanded his experiment to two different types of tiles, one red and one green, with self-complementary edges but incompatible with the other, and shook them. The self-assembly of separate red and green spheres took less than ten minutes.
"Even though we knew that this type of self-organization occurs spontaneously in nature, watching it happen from random shaking was an inspiration," Olson said.
(Films of both these experiments can be viewed at http://www.pnas.org/cgi/content/full/0709489104/DC1)
Virally Inspired Design
From that spark, Olson and his colleagues moved to a closer examination of the assembly principles using synthetic chemistry.
"The question we asked was, 'How can you make something from corannulene core that will behave the way we want it to behave?'" he said. "We built a computational model of the synthetic possibilities to see if they would work if we shook them up. After doing the quantum mechanics and the molecular dynamics, we showed that the process worked in our computer simulation. But this is still very much a theory. Corannulene is a well established molecule but the actual functionalization—creating the right type of stickiness on the edges—has proven to be difficult. We're still working on that."
(An animated presentation of Olson's computer simulation is also available at http://www.pnas.org/cgi/content/full/0709489104/DC1).
After choosing the corannulene-skeleton, the scientists considered three different binding mechanisms—hydrogen bonding, metal binding, and formation of disulfide bonds. Each type, the study said, provides different advantages.
"Each of these bindings has a different characteristic of assembly and disassembly," Olson said. "The various binding choices give you a range of chemistries and environmental conditions with which to assemble and disassemble, a menu of choices to affect the process."
Because visualization is so much a part of his laboratory, Olson is interested in using the physical models as teaching tools. He's shown it to several people and they agree that the model can teach certain fundamental biological functions quite well—even if it seems a bit like magic at first.
"How many times do you see individual objects being shaken together to form a perfectly organized spherical container?" Olson said. "It seems counterintuitive, but it's a real phenomenon."
In addition to Olson, authors of the study are Yunfeng H. E. Hu of The Scripps Research Institute and Ehud Keinan of The Scripps Research Institute and Technion–Israel Institute of Technology, Technion City.
The study, Chemical Mimicry Of Viral Capsid Self-Assembly, was supported by the National Institutes of Health, the National Science Foundation Grant, the Israel Science Foundation, the German–Israeli Project Cooperation (DIP), the Institute of Catalysis Science and Technology, Technion, and The Skaggs Institute for Chemical Biology. See http://www.pnas.org/cgi/content/abstract/104/52/20731.
Send comments to: mikaono[at]scripps.edu
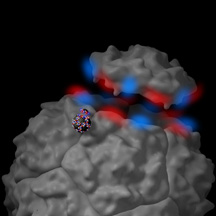
A chemical capsid mimics the capsid assembly of a poliovirus. Click for image details.
