

Metabolite Profiling Rings in New Year
By Jason Socrates Bardi
This is the month where the past comes face-to-face with the future, where we move from boisterous Auld Lang Syne reflections to unrealistic New Year's resolutions. Fittingly, the Romans named the first month of the year January after their god of gates, Janus—the guy with two faces, one looking to the future and one to the past. Like so much else from Roman culture, the month of January has survived to this day.
Janus has demonstrated staying power as well. Modern western culture has made him a symbol for everything from computer servers to retirement funds. At risk of angering the Roman gods, there may still be room for one more metaphor: proteomics, which examines the expression, location, concentration and activity of specific proteins, also has two faces like Janus.
One of the faces of proteomics looks back on the Human Genome Project, and the field is increasing our knowledge of what the enzymes and other proteins expressed by the genes in the human genome do. The other face looks to the future, offering the possibility of discovering how proteins are involved in human health and charting the path forward for finding new ways fight human diseases.
However, that is sometimes easier said than done.
One problem scientists face in proteomics, says Professor Benjamin Cravatt of The Scripps Research Institute, is that many times a proteomic experiment might yield dozens or sometimes hundreds of gene products that are involved in a given disease, but little information about what these proteins are actually doing. In other words, proteomics may uncover that a number of enzymes are involved in one particular disease, but not necessarily how they are involved.
Now a new method for determining the "how"is described in a recent paper in the journal Biochemistry, says Cravatt, who is a professor in the Departments of Cell Biology and Chemistry, director of the Helen L. Dorris Institute for the Study of Neurological and Psychiatric Disorders, and a member of The Skaggs Institute for Chemical Biology at Scripps Research.
Discovery Metabolite Profiling
The question of how enzymes are involved in particular pathologies could be better answered if we knew what these enzymes do biochemically or physiologically, says Cravatt.
There are a number of methods that have emerged in recent years for identifying the physiological functions of enzymes inside cells of the human body. One of the basic approaches, using techniques like RNAi and targeted gene disruption, can tell you what an enzyme does by eliminating it and looking at the effect.
But these techniques do not necessarily yield information on the stepwise biochemical mechanism through which the enzymes act and the substrates the enzymes accepts along the way. These questions, which are of vital importance for understanding how particular enzymes are involved in health and disease, are still addressed mostly through in vitro assays using purified preparations of protein.
Hoping to improve on this, Alan Saghatelian, a postdoctoral fellow in the Cravatt group and a graduate of the Kellogg School of Science and Technology at The Scripps Research Institute has invented a new mass spectrometry-based method that can help sort out the biochemical function of enzymes directly in the tissues where these proteins are natively expressed. In a recent issue of the journal Biochemistry, Saghatelian, Cravatt and their colleagues describe the method, which they refer to as discovery metabolite profiling (DMP).
DMP basically returns a list of an enzyme's endogenous substrates—the natural molecules inside a cell that are regulated by that enzyme. It compares the contents of normal cells to those in which an enzyme in question has been inactivated. By separating the small molecule contents of the cells and looking at every mass in a given range, it is possible to identify the metabolites acted upon by the enzyme.
In their recent paper, Saghatelian, Cravatt and their colleagues applied DMP to a mammalian enzyme Cravatt has studied since he was a graduate student. The enzyme is fatty acid amide hydrolase (FAAH), a membrane-bound enzyme in the brain that metabolizes small molecules, including endocannabinoids—endogenous molecules that provide some natural relief when you feel pain. FAAH is a target for pain therapy because it breaks down the molecules that provide the pain relief.
Significantly, when Saghatelian, Cravatt and their colleagues used DMP to determine the brain lipids regulated by FAAH, they were able to detect both known neural signaling molecules, such as the endocannabinoids, and also an entirely new class of brain-enriched natural products that are regulated by FAAH—that were structurally characterized as taurine-conjugated fatty acids. This suggests FAAH may act as a regulator of neuronal levels of taurine, a key signaling molecule in the brain.
To read the article, "Assignment of Endogenous Substrates to Enzymes by Global Metabolite Profiling" by Alan Saghatelian, Sunia A. Trauger, Elizabeth J. Want, Edward G. Hawkins, Gary Siuzdak, and Benjamin F. Cravatt, see 2004's Volume 43 of the journal Biochemistry (p. 14,332) or go to http://dx.doi.org/10.1021/bi0480335.
Send comments to: jasonb@scripps.edu
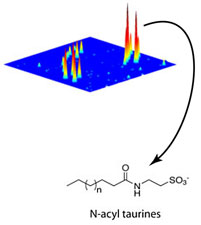
This three-dimensional plot compares the mouse brain metabolomes of FAAH taken from wild-type and FAAH knockout tissues and highlights the dramatic elevation in N-acyl taurines in the knockouts.
