STRUCTURAL MOLECULAR BIOLOGY
AND PROTEIN DESIGN
|
John A. Tainer, Ph.D.
Professor, Department of Molecular Biology
and The Skaggs Institute for Chemical Biology
|
J.
A. Tainer, M. Aoyagi, A. S.
Arvai, J.-L. Bodmer,
D. P. Barondeau, R. M. Brudler,
J.
M. Castagnetto, B. R. Chapados, L. Craig, T. H. Cross, D. S. Daniels,
M.
Didonato, G. DiVita, L. Fan, E.
D. Garcin,
S. W. Hennessy,
K. Hitomi, C. J. Kassman, S. J. Loyd,
C.
D. Mol, G. Moncalian, J.J.
Perry, M. E. Pique, D. S.
Shin, R. S. Williams, M. M. Thayer, T. T. Woo, T. I. Wood
[Recent Graduates] [Former
members]
Other Useful Information: Grant Deadlines,
Synchroton
Deadlines
Key words: DNA-repair, metalloenzyme, oxidative damage, degenerative
disease, infectious disease, Lou Gehrig's disease, superoxide dismutase,
cell cycle, cancer, protein design
 IMMEDIATE
OPENING - SEEKING BIOCHEMIST/MOLECULAR BIOLOGIST.
IMMEDIATE
OPENING - SEEKING BIOCHEMIST/MOLECULAR BIOLOGIST. 
 SEEKING
MACROMOLECULAR CRYSTALLOGRAPHERS - Positions Available.
SEEKING
MACROMOLECULAR CRYSTALLOGRAPHERS - Positions Available. 
Visit some of our collaborators at TSRI: [E.
D. Getzoff] [V. A.
Roberts][Metalloprotein
Structure and Design Group]
Internal Pages: [Beamline
Phone Numbers]
 AP Endonuclease DNA repair enzyme
AP Endonuclease DNA repair enzyme
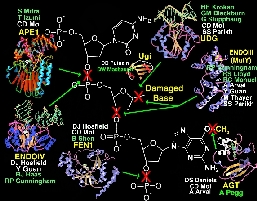 DNA Damage Excision Enzyme Structures
DNA Damage Excision Enzyme Structures
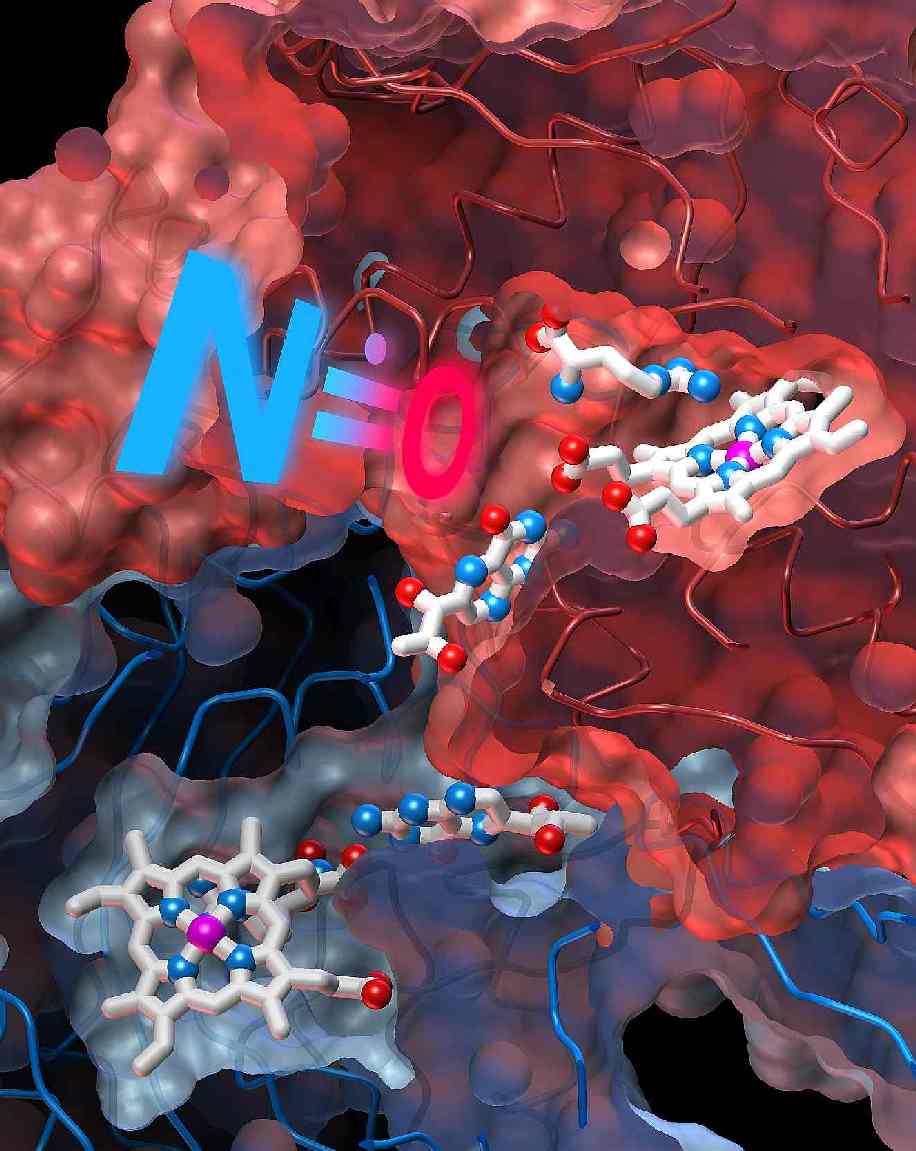
Structure of Nitric Oxide Synthase
Oxygenase Dimer with Pterin and Substrate.
B.R. Crane, A.S. Arvai, D.K.
Ghosh, C. Wu, E.D. Getzoff, D.J. Stuehr and J.A. Tainer.
Science 279, 2121-21216,
1998. Visit Science
Magazine Online
Check out our NOS
Steroeviews and Figures.
We focus on four classes of proteins that interface structural biology
with cellular chemistry:
-
Enzymes regulating reactive oxygen and xenotoxin control (superoxide dismutases,
catalase, and glutathione transferase);
-
Enzymes controlling DNA repair and evolution enzymes;
-
Pilus fiber motility and adherence proteins for bacterial pathogens;
-
Cell cycle control proteins.
Current structural and design results are contributing to an understanding
of these systems that should contribute to new treatments for infectious
disease, degenerative diseases, and cancer.
Bacterial pilus structure, assembly, and antigenicity.
We seek to improve understanding and to develop new treatments for the
many bacterial pathogens that use long fibers termed Type IV pili for attachment
and mobility. We solved the crystallographic structure of the pilus fiber-
forming pilin protein essential to the virulence of these pathogens, which
includes Neisseria meningitidis, Neisseria gonorrhoeae, Pseudomonas aeroginosa,
Dichlobacter nodosus, Moraxella bovis, Vibrio cholerae, and enterotoxogenic
Escherichia coli. The pilin subunit is ladle- shaped with an extended a-helix
spine wrapped at one end by b-sheet. A few key
residue interactions allow the remaining part of a hypervariable region
to undergo extreme antigenic variation to escape t he host immune response.
The assembled fiber shows extreme sequence variation plus glycosylation
and phosphorylation at the survace. However, we have defined two conserved
regions recognized by human, mouse, and rabbit antibodies. Current efforts
include the redesign of pilin to develop potential vaccines by replacing
the hypervariable region with epitopes from other essential target proteins.
Reactive oxygen and xenobiotic control enzymes.
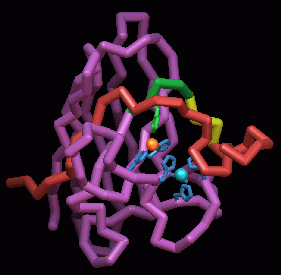
Atomic structures of human cytoplasmic Cu,Zn superoxide dismutases,
the mitochondrial Mn superoxide dismutases, and schistosomal glutathione
transferases are improving our understanding of reactive oxygen and xenobiotic
control within cells. Superoxide dismutases (SODs) are master regulators
for reactive oxygen species involved in injury, pathogenesis, aging and
degenerative diseases. For Cu,Zn SOD, we have defined the active site structural
chemistry responsible for the rapid reaction. We are now exam ining how
single site mutations cause degenerative disease such as Lou Gehrig's disease
or familial amyotrophic lateral sclerosis (FALS). For Mn SOD, single site
mutations can destabilize the tetramer and also reduce stability and activity
in ways that ma y cause degenerative diseases. Structures of glutathione-S-transferase
(GST), which is an essential detoxification enzyme in all organisms, show
how the leading anti-schistosomal drug praziquantel binds to GST. This
information may allow the design of new drugs to overcome the growing resistance
to current schistosomal drugs. Protein design on GST enzymes includes using
random libraries for the loop regions surrounding the active site, thereby
developing glubodies as a new class of binding proteins in bio technology.
DNA-repair and genetic evolution.
 Cells must balance DNA repair
to preserve fidelity with DNA variation allowing evolutionary changes.
As over 10,000 DNA bases per day are repaired in each human cell, DNA excision-repair
enzymes are essential to cell survival and to protection against cancer-causing
mutations. Surprisingly, DNA repair inhibitors may improve current radiation
and chemotherapies for cancer by specifically killing cancer cells which
unlike normal cells will often undergo DNA synthesis and cell division
with unrepaired DNA resulting in their death. Our DNA repair enzyme structures
show how damaged DNA bases are recognized and removed in atomic detail.
These enzymes repair DNA by flipping the DNA nucleotides out from the double
helix and into specific binding pockets (Figure), which are ideal for the
design of inhibitors for anti-cancer therapies. We confirmed our understanding
of these binding pockets by deliberately altering the specificity of the
DNA repair enzyme uracil-DNA glycosylase to remove cytosine or thymine
from normal DNA resulting in mutator phenotypes in vivo. We found that
the endonuclease III structure is representative of a superfamily of DNA
repair enzymes and a key HhH motif that recognizes DNA backbone. Our new
structure of the major DNA-repair APendonuclease, which cuts DNA at sites
where bases are missing, defines its active site and identifies mechanism
for recognizing missing bases.
Cells must balance DNA repair
to preserve fidelity with DNA variation allowing evolutionary changes.
As over 10,000 DNA bases per day are repaired in each human cell, DNA excision-repair
enzymes are essential to cell survival and to protection against cancer-causing
mutations. Surprisingly, DNA repair inhibitors may improve current radiation
and chemotherapies for cancer by specifically killing cancer cells which
unlike normal cells will often undergo DNA synthesis and cell division
with unrepaired DNA resulting in their death. Our DNA repair enzyme structures
show how damaged DNA bases are recognized and removed in atomic detail.
These enzymes repair DNA by flipping the DNA nucleotides out from the double
helix and into specific binding pockets (Figure), which are ideal for the
design of inhibitors for anti-cancer therapies. We confirmed our understanding
of these binding pockets by deliberately altering the specificity of the
DNA repair enzyme uracil-DNA glycosylase to remove cytosine or thymine
from normal DNA resulting in mutator phenotypes in vivo. We found that
the endonuclease III structure is representative of a superfamily of DNA
repair enzymes and a key HhH motif that recognizes DNA backbone. Our new
structure of the major DNA-repair APendonuclease, which cuts DNA at sites
where bases are missing, defines its active site and identifies mechanism
for recognizing missing bases.
Human dUTP pyrophosphatase (dUTPase) catalyzes the breakdown of uracil
nucleotide triphosphates to keep the RNA base uracil out of DNA and to
provide material for the biosynthesis of the DNA building block dTTP. These
dUTPase functions prevent thymic-less cell death from cycles of uracil
misincorporation and removal that would generate multiple DNA strand breaks
and eventual cell death. Atomic structures of dUTPase with bound nucleotides
reveal uracil binds within a groove that is then capped when the flexible
tail region closes over the bound dUTP substrate. These structures establish
how dUTPase recognizes its substrate with exquisite specificity and provide
a basis for the design of inhibitors as future anti-cancer drugs.
Cell cycle control.
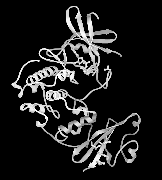 Together with Steve Reed's
group, we are working to define structural controls on cell cycle progression.
Structures of the Cks or suc1 proteins which are essential to cell cycle
progression, provide clues for new mechanisms for cell cycle regulation
via a conformational switch that controls two distinct Cks folds and assemblies.
A straight b-hinge conformation of Cks, which
forms a dimer of swapped b-strands blocks binding
to the cell cycle kinase Cdk2. Formation of a closed, bent b-
hinge conformation creates a single domain fold that promotes Cdk2 binding
(Figure). Preliminary experiments by Steve Reed's group show that blocking
Cks expression results in cell death for several types of cancer cells
suggesting this is a useful cancer drug target.
Together with Steve Reed's
group, we are working to define structural controls on cell cycle progression.
Structures of the Cks or suc1 proteins which are essential to cell cycle
progression, provide clues for new mechanisms for cell cycle regulation
via a conformational switch that controls two distinct Cks folds and assemblies.
A straight b-hinge conformation of Cks, which
forms a dimer of swapped b-strands blocks binding
to the cell cycle kinase Cdk2. Formation of a closed, bent b-
hinge conformation creates a single domain fold that promotes Cdk2 binding
(Figure). Preliminary experiments by Steve Reed's group show that blocking
Cks expression results in cell death for several types of cancer cells
suggesting this is a useful cancer drug target.
Publications
-
Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA Structure
and mechanism of the RuvB Holliday junction branch migrati J Mol Biol.
2001 Aug 10;311(2):297-310.
-
Chung JH, Suh MJ, Park YI, Tainer JA, Han YS. Repair activities of 8-oxoguanine
DNA glycosylase from Archaeoglobus hyperthermophilic archaeon. Mutat Res.
2001 Jul 12;486(2):99-111.
-
Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural
biochemistry and interaction architecture of the DNA doub break repair
Mre11 nuclease and Rad50-ATPase. Cell. 2001 May 18;105(4):473-85.
-
Ayala BP, Vasquez B, Clary S, Tainer JA, Rodland K, So M. The pilus-induced
Ca2+ flux triggers lysosome exocytosis and increas amount of Lamp1 accessible
to Neisseria IgA1 protease. Cell Microbiol. 2001 Apr;3(4):265-75.
-
Chapados BR, Chai Q, Hosfield DJ, Qiu J, Shen B, Tainer JA. Structural
biochemistry of a type 2 RNase H: RNA primer recognition during DNA replication.
J Mol Biol. 2001 Mar 23;307(2):541-56.
-
Hopfner KP, Tainer JA. DNA mismatch repair: the hands of a genome guardian.
Structure Fold Des. 2000 Dec 15;8(12):R237-41. Review.
-
Han S, Arvai AS, Clancy SB, Tainer JA. Crystal structure and novel recognition
motif of rho ADP-ribosylatin exoenzyme from Clostridium botulinum: structural
insights for recogn specificity and catalysis. J Mol Biol. 2001 Jan 5;305(1):95-107.
-
Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP. Mre11 and
Rad50 from Pyrococcus furiosus: cloning and biochemical characterization
reveal an evolutionarily conserved multiprotein mac J Bacteriol. 2000 Nov;182(21):6036-41.
-
Bourne Y, Watson MH, Arvai AS, Bernstein SL, Reed SI, Tainer JA. Crystal
structure and mutational analysis of the Saccharomyces cerev cycle regulatory
protein Cks1: implications for domain swapping, ani and protein interactions.
Structure Fold Des. 2000 Aug 15;8(8):841-50.
-
Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic
endonuclease fa DNA base excision repair: the 3' ends justify the means.
Mutat Res. 2000 Aug 30;460(3-4):211-29. Review.
-
Parikh SS, Putnam CD, Tainer JA. Lessons learned from structural results
on uracil-DNA glycosylase. Mutat Res. 2000 Aug 30;460(3-4):183-99. Review.
-
Daniels DS, Tainer JA. Conserved structural motifs governing the stoichiometric
repair of a by O(6)-alkylguanine-DNA alkyltransferase. Mutat Res. 2000
Aug 30;460(3-4):151-63. Review.
-
Tainer JA, Friedberg EC. Dancing with the elephants: envisioning the structural
biology of DN pathways. Mutat Res. 2000 Aug 30;460(3-4):139-41. No abstract
available.
-
Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, T Structural
biology of Rad50 ATPase: ATP-driven conformational contro double-strand
break repair and the ABC-ATPase superfamily. Cell. 2000 Jun 23;101(7):789-800.
-
Leveque VJ, Stroupe ME, Lepock JR, Cabelli DE, Tainer JA, Nick H DN. Multiple
replacements of glutamine 143 in human manganese superoxide effects on
structure, stability, and catalysis. Biochemistry. 2000 Jun 20;39(24):7131-7.
-
Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackbur JA. Uracil-DNA
glycosylase-DNA substrate and product structures: conform strain promotes
catalytic efficiency by coupled stereoelectronic eff Proc Natl Acad Sci
U S A. 2000 May 9;97(10):5083-8.
-
Koc ON, Gerson SL, Cooper BW, Laughlin M, Meyerson H, Kutteh L, Szekely
EM, Tainer N, Lazarus HM. Randomized cross-over trial of progenitor-cell
mobilization: high-do cyclophosphamide plus granulocyte colony-stimulating
factor (G-CSF) granulocyte-macrophage colony-stimulating factor plus G-CSF.
J Clin Oncol. 2000 May;18(9):1824-30.
-
Crane BR, Arvai AS, Ghosh S, Getzoff ED, Stuehr DJ, Tainer JA. Structures
of the N(omega)-hydroxy-L-arginine complex of inducible n synthase oxygenase
dimer with active and inactive pterins. Biochemistry. 2000 Apr 25;39(16):4608-21.
-
Daniels DS, Mol CD, Arvai AS, Kanugula S, Pegg AE, Tainer JA. Active and
alkylated human AGT structures: a novel zinc site, inhibi extrahelical
base binding. EMBO J. 2000 Apr 3;19(7):1719-30.
-
Zu JS, Deng HX, Lo TP, Mitsumoto H, Ahmed MS, Hung WY, Cai ZJ, T Siddique
T. Exon 5 encoded domain is not required for the toxic function of muta
essential for the dismutase activity: identification and characteriz new
SOD1 mutations associated with familial amyotrophic lateral scle Neurogenetics.
1997 May;1(1):65-71.
-
Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal
abasic DNA binding by APE1 a repair coordination [corrected] Nature. 2000
Jan 27;403(6768):451-6.
-
Putnam CD, Arvai AS, Bourne Y, Tainer JA. Active and inhibited human catalase
structures: ligand and NADPH bin catalytic mechanism. J Mol Biol. 2000
Feb 11;296(1):295-309.
-
Putnam CD, Tainer JA. The food of sweet and bitter fancy. Nat Struct Biol.
2000 Jan;7(1):17-8.
-
Crane BR, Rosenfeld RJ, Arvai AS, Ghosh DK, Ghosh S, Tainer JA, Getzoff
ED. N-terminal domain swapping and metal ion binding in nitric oxide syn
dimerization. EMBO J. 1999 Nov 15;18(22):6271-81.
-
Ghosh DK, Crane BR, Ghosh S, Wolan D, Gachhui R, Crooks C, Prest JA, Getzoff
ED, Stuehr DJ. Inducible nitric oxide synthase: role of the N-terminal
beta-hairpin pterin-binding segment in dimerization and tetrahydrobiopterin
inter EMBO J. 1999 Nov 15;18(22):6260-70.
-
Cabelli DE, Guan Y, Leveque V, Hearn AS, Tainer JA, Nick HS, Sil Role of
tryptophan 161 in catalysis by human manganese superoxide di Biochemistry.
1999 Sep 7;38(36):11686-92.
-
Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. Evolution and mechanism
from structures of an ADP-ribosylating toxin complex. Nat Struct Biol.
1999 Oct;6(10):932-6.
-
Ramilo CA, Leveque V, Guan Y, Lepock JR, Tainer JA, Nick HS, Sil Interrupting
the hydrogen bond network at the active site of human m superoxide dismutase.
J Biol Chem. 1999 Sep 24;274(39):27711-6.
-
Adak S, Crooks C, Wang Q, Crane BR, Tainer JA, Getzoff ED, Stueh Tryptophan
409 controls the activity of neuronal nitric-oxide syntha regulating nitric
oxide feedback inhibition. J Biol Chem. 1999 Sep 17;274(38):26907-11.
-
Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the
DNA repair enzyme endonuclease IV and its DNA compl double-nucleotide flipping
at abasic sites and three-metal-ion catal Cell. 1999 Aug 6;98(3):397-408.
-
Ghosh S, Wolan D, Adak S, Crane BR, Kwon NS, Tainer JA, Getzoff DJ. Mutational
analysis of the tetrahydrobiopterin-binding site in induc nitric-oxide
synthase. J Biol Chem. 1999 Aug 20;274(34):24100-12.
-
Pellequer JL, Chen Sw, Roberts VA, Tainer JA, Getzoff ED. Unraveling the
effect of changes in conformation and compactness at V(L)-V(H) interface
upon antigen binding. J Mol Recognit. 1999 Jul-Aug;12(4):267-75.
-
Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for
the recognition and removal of damaged DNA Annu Rev Biophys Biomol Struct.
1999;28:101-28. Review.
-
Bruns CM, Hubatsch I, Ridderstrom M, Mannervik B, Tainer JA. Human glutathione
transferase A4-4 crystal structures and mutagenesi basis of high catalytic
efficiency with toxic lipid peroxidation pro J Mol Biol. 1999 May 7;288(3):427-39.
-
Haas BJ, Sandigursky M, Tainer JA, Franklin WA, Cunningham RP. Purification
and characterization of Thermotoga maritima endonucleas thermostable apurinic/apyrimidinic
endonuclease and 3'-repair dieste J Bacteriol. 1999 May;181(9):2834-9.
-
Shroyer MJ, Bennett SE, Putnam CD, Tainer JA, Mosbaugh DW. Mutation of
an active site residue in Escherichia coli uracil-DNA gl effect on DNA
binding, uracil inhibition and catalysis. Biochemistry. 1999 Apr 13;38(15):4834-45.
-
Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh JA. Protein
mimicry of DNA from crystal structures of the uracil-DNA gly inhibitor
protein and its complex with Escherichia coli uracil-DNA g J Mol Biol.
1999 Mar 26;287(2):331-46.
-
Forest KT, Dunham SA, Koomey M, Tainer JA. Crystallographic structure reveals
phosphorylated pilin from Neisser phosphoserine sites modify type IV pilus
surface chemistry and fibre Mol Microbiol. 1999 Feb;31(3):743-52.
-
Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography
of DNA base excision repair. Curr Opin Struct Biol. 1999 Feb;9(1):37-47.
Review.
-
Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd JA. MutY
catalytic core, mutant and bound adenine structures define spec DNA repair
enzyme superfamily. Nat Struct Biol. 1998 Dec;5(12):1058-64.
-
Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and
replication endonuclease and exonucl coupling DNA and PCNA binding to FEN-1
activity. Cell. 1998 Oct 2;95(1):135-46.
-
Hosfield DJ, Frank G, Weng Y, Tainer JA, Shen B. Newly discovered archaebacterial
flap endonucleases show a structure mechanism for DNA substrate binding
and catalysis resembling human f endonuclease-1. J Biol Chem. 1998 Oct
16;273(42):27154-61.
-
Coombs GS, Bergstrom RC, Pellequer JL, Baker SI, Navre M, Smith JA, Madison
EL, Corey DR. Substrate specificity of prostate-specific antigen (PSA).
Chem Biol. 1998 Sep;5(9):475-88.
-
Schnecke V, Swanson CA, Getzoff ED, Tainer JA, Kuhn LA. Screening a peptidyl
database for potential ligands to proteins with flexibility. Proteins.
1998 Oct 1;33(1):74-87.
-
Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision
repair initiation revealed by crystal structures and b kinetics of human
uracil-DNA glycosylase with DNA. EMBO J. 1998 Sep 1;17(17):5214-26.
-
Shen B, Qiu J, Hosfield D, Tainer JA. Flap endonuclease homologs in archaebacteria
exist as independent pr Trends Biochem Sci. 1998 May;23(5):171-3. Review.
No abstract availa
-
Tong W, Burdi D, Riggs-Gelasco P, Chen S, Edmondson D, Huynh BH, Han S,
Arvai A, Tainer J. Characterization of Y122F R2 of Escherichia coli ribonucleotide
redu time-resolved physical biochemical methods and X-ray crystallography
Biochemistry. 1998 Apr 28;37(17):5840-8.
-
Hsieh Y, Guan Y, Tu C, Bratt PJ, Angerhofer A, Lepock JR, Hickey JA, Nick
HS, Silverman DN. Probing the active site of human manganese superoxide
dismutase: the glutamine 143. Biochemistry. 1998 Apr 7;37(14):4731-9.
-
Guan Y, Hickey MJ, Borgstahl GE, Hallewell RA, Lepock JR, O'Conn Y, Nick
HS, Silverman DN, Tainer JA. Crystal structure of Y34F mutant human mitochondrial
manganese super dismutase and the functional role of tyrosine 34. Biochemistry.
1998 Apr 7;37(14):4722-30.
-
Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Taine Structure
of nitric oxide synthase oxygenase dimer with pterin and s Science. 1998
Mar 27;279(5359):2121-6.
-
Marceau M, Forest K, Beretti JL, Tainer J, Nassif X. Consequences of the
loss of O-linked glycosylation of meningococcal pilin on piliation and
pilus-mediated adhesion. Mol Microbiol. 1998 Feb;27(4):705-15.
-
Koc ON, Gerson SL, Phillips GL, Cooper BW, Kutteh L, Van Zant G, Fox RM,
Schupp JE, Tainer N, Lazarus HM. Autologous CD34+ cell transplantation
for patients with advanced lym effects of overnight storage on peripheral
blood progenitor cell enr engraftment. Bone Marrow Transplant. 1998 Feb;21(4):337-43.
-
Parikh SS, Mol CD, Tainer JA. Base excision repair enzyme family portrait:
integrating the structu chemistry of an entire DNA repair pathway. Structure.
1997 Dec 15;5(12):1543-50. Review.
-
Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tain Hickson
ID, Freemont PS. The crystal structure of the human DNA repair endonuclease
HAP1 sugg recognition of extra-helical deoxyribose at DNA abasic sites.
EMBO J. 1997 Nov 3;16(21):6548-58.
-
Crane BR, Arvai AS, Gachhui R, Wu C, Ghosh DK, Getzoff ED, Stueh JA. The
structure of nitric oxide synthase oxygenase domain and inhibito Science.
1997 Oct 17;278(5337):425-31.
-
Fisher CL, Cabelli DE, Hallewell RA, Beroza P, Lo TP, Getzoff ED Computational,
pulse-radiolytic, and structural investigations of ly its role in the electrostatic
triad of human Cu,Zn superoxide dismut Proteins. 1997 Sep;29(1):103-12.
-
Forest KT, Tainer JA. Type-4 pilus-structure: outside to inside and top
to bottom--a minir Gene. 1997 Jun 11;192(1):165-9. Review.
-
Boissinot M, Karnas S, Lepock JR, Cabelli DE, Tainer JA, Getzoff Hallewell
RA. Function of the Greek key connection analysed using circular permuta
superoxide dismutase. EMBO J. 1997 May 1;16(9):2171-8.
-
Roberts VA, Nachman RJ, Coast GM, Hariharan M, Chung JS, Holman H, Tainer
JA. Consensus chemistry and beta-turn conformation of the active core of
kinin neuropeptide family. Chem Biol. 1997 Feb;4(2):105-17.
-
Bourne Y, Redford SM, Steinman HM, Lepock JR, Tainer JA, Getzoff Novel
dimeric interface and electrostatic recognition in bacterial C superoxide
dismutase. Proc Natl Acad Sci U S A. 1996 Nov 12;93(23):12774-9.
-
Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping
mechanism from the structure of human uracil-D glycosylase bound to DNA.
Nature. 1996 Nov 7;384(6604):87-92.
-
Mol CD, Harris JM, McIntosh EM, Tainer JA. Human dUTP pyrophosphatase:
uracil recognition by a beta hairpin and sites formed by three separate
subunits. Structure. 1996 Sep 15;4(9):1077-92.
-
Watson MH, Bourne Y, Arvai AS, Hickey MJ, Santiago A, Bernstein JA, Reed
SI. A mutation in the human cyclin-dependent kinase interacting protein,
interferes with cyclin-dependent kinase binding and biological funct preserves
protein structure and assembly. J Mol Biol. 1996 Sep 6;261(5):646-57.
-
Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Excision
of cytosine and thymine from DNA by mutants of human uracil glycosylase.
EMBO J. 1996 Jul 1;15(13):3442-7.
-
Napolitano EW, Villar HO, Kauvar LM, Higgins DL, Roberts D, Mand SK, Engqvist-Goldstein
A, Bukar R, Calio BL, Jack HM, Tainer JA. Glubodies: randomized libraries
of glutathione transferase enzymes. Chem Biol. 1996 May;3(5):359-67.
-
Borgstahl GE, Parge HE, Hickey MJ, Johnson MJ, Boissinot M, Hall Lepock
JR, Cabelli DE, Tainer JA. Human mitochondrial manganese superoxide dismutase
polymorphic varia reduces activity by destabilizing the tetrameric interface.
Biochemistry. 1996 Apr 9;35(14):4287-97.
-
Bourne Y, Watson MH, Hickey MJ, Holmes W, Rocque W, Reed SI, Tai Crystal
structure and mutational analysis of the human CDK2 kinase c cell cycle-regulatory
protein CksHs1. Cell. 1996 Mar 22;84(6):863-74.
-
Ogihara NL, Parge HE, Hart PJ, Weiss MS, Goto JJ, Crane BR, Tsan K, Roe
JA, Valentine JS, Eisenberg D, Tainer JA. Unusual trigonal-planar copper
configuration revealed in the atomic yeast copper-zinc superoxide dismutase.
Biochemistry. 1996 Feb 20;35(7):2316-21.
-
Forest KT, Bernstein SL, Getzoff ED, So M, Tribbick G, Geysen HM Tainer
JA. Assembly and antigenicity of the Neisseria gonorrhoeae pilus mapped
antibodies. Infect Immun. 1996 Feb;64(2):644-52.
-
Kuhn LA, Swanson CA, Pique ME, Tainer JA, Getzoff ED. Atomic and residue
hydrophilicity in the context of folded protein s Proteins. 1995 Dec;23(4):536-47.
-
Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tain Structure
of the fibre-forming protein pilin at 2.6 A resolution. Nature. 1995 Nov
2;378(6552):32-8.
-
Bourne Y, Arvai AS, Bernstein SL, Watson MH, Reed SI, Endicott J Johnson
LN, Tainer JA. Crystal structure of the cell cycle-regulatory protein suc1
reveals conformational switch. Proc Natl Acad Sci U S A. 1995 Oct 24;92(22):10232-6.
-
Phillips JP, Tainer JA, Getzoff ED, Boulianne GL, Kirby K, Hilli Subunit-destabilizing
mutations in Drosophila copper/zinc superoxide neuropathology and a model
of dimer dysequilibrium. Proc Natl Acad Sci U S A. 1995 Sep 12;92(19):8574-8.
-
Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, DW, Tainer
JA. Crystal structure of human uracil-DNA glycosylase in complex with a
inhibitor: protein mimicry of DNA. Cell. 1995 Sep 8;82(5):701-8.
-
Pond L, Kuhn LA, Teyton L, Schutze MP, Tainer JA, Jackson MR, Pe A role
for acidic residues in di-leucine motif-based targeting to th pathway.
J Biol Chem. 1995 Aug 25;270(34):19989-97.
-
Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding
motifs in the DNA repair enzyme endonuclease III c structure. EMBO J. 1995
Aug 15;14(16):4108-20.
-
Barzilay G, Mol CD, Robson CN, Walker LJ, Cunningham RP, Tainer ID. Identification
of critical active-site residues in the multifunction repair enzyme HAP1.
Nat Struct Biol. 1995 Jul;2(7):561-8.
-
Arvai AS, Bourne Y, Hickey MJ, Tainer JA. Crystal structure of the human
cell cycle protein CksHs1: single dom with similarity to kinase N-lobe
domain. J Mol Biol. 1995 Jun 23;249(5):835-42.
-
Deng HX, Tainer JA, Mitsumoto H, Ohnishi A, He X, Hung WY, Zhao Hentati
A, Siddique T. Two novel SOD1 mutations in patients with familial amyotrophic
later sclerosis. Hum Mol Genet. 1995 Jun;4(6):1113-6. No abstract available.
-
Sjalander A, Beckman G, Deng HX, Iqbal Z, Tainer JA, Siddique T. The D90A
mutation results in a polymorphism of Cu,Zn superoxide dism is prevalent
in northern Sweden and Finland. Hum Mol Genet. 1995 Jun;4(6):1105-8. No
abstract available.
-
McTigue MA, Bernstein SL, Williams DR, Tainer JA. Purification and crystallization
of a schistosomal glutathione S-tra Proteins. 1995 May;22(1):55-7.
-
Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tai Crystal
structure and mutational analysis of human uracil-DNA glycos structural
basis for specificity and catalysis. Cell. 1995 Mar 24;80(6):869-78.
-
Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function
of the multifunctional DNA-repair enzyme exon Nature. 1995 Mar 23;374(6520):381-6.
-
McTigue MA, Williams DR, Tainer JA. Crystal structures of a schistosomal
drug and vaccine target: glutat S-transferase from Schistosoma japonica
and its complex with the lea antischistosomal drug praziquantel. J Mol
Biol. 1995 Feb 10;246(1):21-7.
-
Tainer JA, Thayer MM, Cunningham RP. DNA repair proteins. Curr Opin Struct
Biol. 1995 Feb;5(1):20-6. Review.
-
Arvai AS, Bourne Y, Williams D, Reed SI, Tainer JA. Crystallization and
preliminary crystallographic study of human CksH cycle regulatory protein.
Proteins. 1995 Jan;21(1):70-3.
Recent TSRI Graduates from the Tainer Lab
Former members of the Tainer Lab
![]() IMMEDIATE
OPENING - SEEKING BIOCHEMIST/MOLECULAR BIOLOGIST.
IMMEDIATE
OPENING - SEEKING BIOCHEMIST/MOLECULAR BIOLOGIST. ![]()
![]() SEEKING
MACROMOLECULAR CRYSTALLOGRAPHERS - Positions Available.
SEEKING
MACROMOLECULAR CRYSTALLOGRAPHERS - Positions Available. ![]()
 AP Endonuclease DNA repair enzyme
AP Endonuclease DNA repair enzyme DNA Damage Excision Enzyme Structures
DNA Damage Excision Enzyme Structures

 Cells must balance DNA repair
to preserve fidelity with DNA variation allowing evolutionary changes.
As over 10,000 DNA bases per day are repaired in each human cell, DNA excision-repair
enzymes are essential to cell survival and to protection against cancer-causing
mutations. Surprisingly, DNA repair inhibitors may improve current radiation
and chemotherapies for cancer by specifically killing cancer cells which
unlike normal cells will often undergo DNA synthesis and cell division
with unrepaired DNA resulting in their death. Our DNA repair enzyme structures
show how damaged DNA bases are recognized and removed in atomic detail.
These enzymes repair DNA by flipping the DNA nucleotides out from the double
helix and into specific binding pockets (Figure), which are ideal for the
design of inhibitors for anti-cancer therapies. We confirmed our understanding
of these binding pockets by deliberately altering the specificity of the
DNA repair enzyme uracil-DNA glycosylase to remove cytosine or thymine
from normal DNA resulting in mutator phenotypes in vivo. We found that
the endonuclease III structure is representative of a superfamily of DNA
repair enzymes and a key HhH motif that recognizes DNA backbone. Our new
structure of the major DNA-repair APendonuclease, which cuts DNA at sites
where bases are missing, defines its active site and identifies mechanism
for recognizing missing bases.
Cells must balance DNA repair
to preserve fidelity with DNA variation allowing evolutionary changes.
As over 10,000 DNA bases per day are repaired in each human cell, DNA excision-repair
enzymes are essential to cell survival and to protection against cancer-causing
mutations. Surprisingly, DNA repair inhibitors may improve current radiation
and chemotherapies for cancer by specifically killing cancer cells which
unlike normal cells will often undergo DNA synthesis and cell division
with unrepaired DNA resulting in their death. Our DNA repair enzyme structures
show how damaged DNA bases are recognized and removed in atomic detail.
These enzymes repair DNA by flipping the DNA nucleotides out from the double
helix and into specific binding pockets (Figure), which are ideal for the
design of inhibitors for anti-cancer therapies. We confirmed our understanding
of these binding pockets by deliberately altering the specificity of the
DNA repair enzyme uracil-DNA glycosylase to remove cytosine or thymine
from normal DNA resulting in mutator phenotypes in vivo. We found that
the endonuclease III structure is representative of a superfamily of DNA
repair enzymes and a key HhH motif that recognizes DNA backbone. Our new
structure of the major DNA-repair APendonuclease, which cuts DNA at sites
where bases are missing, defines its active site and identifies mechanism
for recognizing missing bases.
 Together with Steve Reed's
group, we are working to define structural controls on cell cycle progression.
Structures of the Cks or suc1 proteins which are essential to cell cycle
progression, provide clues for new mechanisms for cell cycle regulation
via a conformational switch that controls two distinct Cks folds and assemblies.
A straight b-hinge conformation of Cks, which
forms a dimer of swapped b-strands blocks binding
to the cell cycle kinase Cdk2. Formation of a closed, bent b-
hinge conformation creates a single domain fold that promotes Cdk2 binding
(Figure). Preliminary experiments by Steve Reed's group show that blocking
Cks expression results in cell death for several types of cancer cells
suggesting this is a useful cancer drug target.
Together with Steve Reed's
group, we are working to define structural controls on cell cycle progression.
Structures of the Cks or suc1 proteins which are essential to cell cycle
progression, provide clues for new mechanisms for cell cycle regulation
via a conformational switch that controls two distinct Cks folds and assemblies.
A straight b-hinge conformation of Cks, which
forms a dimer of swapped b-strands blocks binding
to the cell cycle kinase Cdk2. Formation of a closed, bent b-
hinge conformation creates a single domain fold that promotes Cdk2 binding
(Figure). Preliminary experiments by Steve Reed's group show that blocking
Cks expression results in cell death for several types of cancer cells
suggesting this is a useful cancer drug target.