HIV INTEGRASE

In 2004, our computational docking located the viral DNA binding site on
the catalyic domain (CCD) of HIV-IN.
We docked a B-form viral DNA model to a molecular dynamics model of the CCD
(Adesokan et al., 2004) and compared it (Roberts et al., 2013)
with the structure of the prototype foamy virus (PFV) integrase
[S. Hare et al., (2010) Nature, 464, 232-236.
This structure, published in 2009, was the first structure of any
integrase with bound DNA.
The best-energy docked DNA placement (blue phosphate backbone)
is representative of our largest favorable-energy cluster.
One strand of the docked DNA shows good alignment with
the active-site strand of the crystallographic DNA (orange with
red 3'-end) bound to PFV integrase, but it also
penetrates into the active site loop (black) of the PFV CCD.
In the HIV CCD model, this loop corresponds to residues 139-153 (magenta).
The incorrect conformation of this modeled active site loop fills
the groove occupied by unprocessed 5'-end of the viral DNA,
preventing correct positioning of the docked DNA at the active site.
The two metal ions (magenta spheres) of the HIV-1 CCD model show good agreement
with those of the PFV CCD (black spheres).
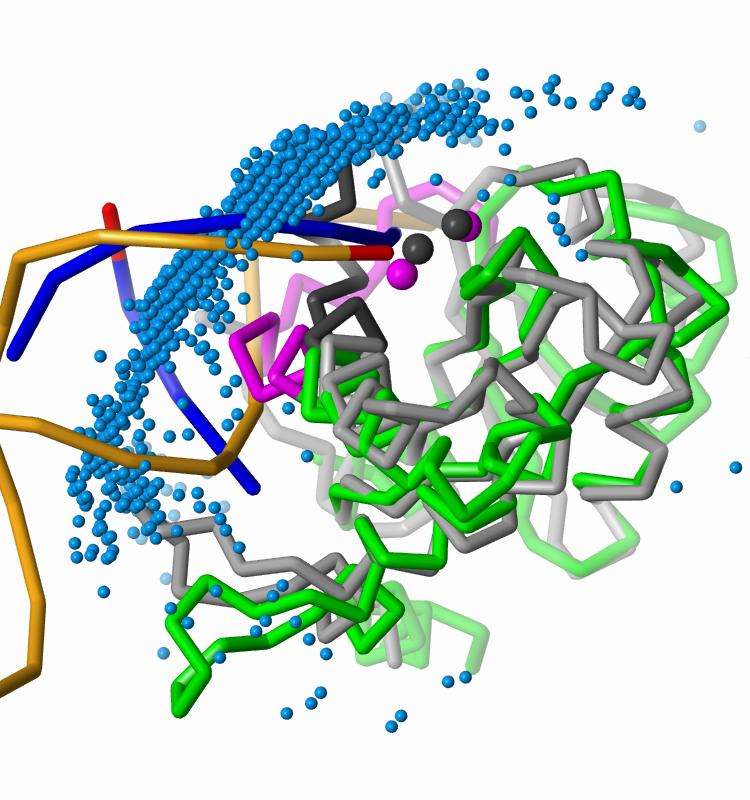
The docked DNA fragments follow the axis of the crystallographic viral DNA.
The 2,000 most favorable docked DNA placements, represented by their
geometric centers (blue spheres), form a large cluster over
the CCD active site and extend over the full surface contacted by
crystallographic viral DNA.

A second, less populated docked DNA cluster
is close to the host DNA binding site found in PFV integrase crystallographic
structures.
The docked DNA (green and light blue)
overlaps the host DNA (yellow, bound to PFV integrase) and
is within 4 A of
the two vDNA 3' processed ends (red) in the PFV integrase tetramer.
Publications
- Adesokan, A. A., Roberts, V. A., Lee, K. W., Lins, R. D., and
Briggs, J. M., (2003)
"Prediction of HIV-1 Integrase/Viral DNA Interactions in the Catalytic
Domain by Fast Molecular Docking"
J. Med. Chem., 47, 821-828.
- Roberts, V. A., Pique, M. E., Ten Eyck, L. F., Li, S., (2013)
"Predicting protein-DNA interactions by full search
computational docking",
Proteins, 81, 2106-2118.
- Roberts, V. A., (2015)
"C-terminal domain of integrase binds between the two active sites",
J. Chem. Theory Comput., 11, 4500-4511.



