

Revealing the Cell Cycle's Secrets
By Jeff Worley
"They're complex, they're sophisticated, and they're marvelously flexible. In their ability to adapt to what can go wrong, they're brilliant really."
Clare McGowan, an associate professor in the Department of Molecular Biology at The Scripps Research Institute, could be talking about anything from a team of arctic researchers to a group of microsurgeons. In fact, she's talking about human cells. Her fascination with human cells—specifically, with how they duplicate—began at the University of Dundee in Scotland, where she earned a Ph.D. in 1987. This interest intensified when she came to Scripps Research in La Jolla, California, the following year.
The Path Less Traveled
In the early 1990s, human cell cycle control was an emerging area of interest and little work had been done on it. But a few discoveries were opening the path for researchers to start looking at these processes in human cells.
"That's when I entered the field," McGowan says. "It was clear even then what we discovered about how the cell cycle works in human cells would be absolutely important to understanding not just how cancer occurs but how we might treat it."
Anticancer therapy is largely based on the use of agents that damage DNA and thus kill dividing cells. So a detailed understanding of cellular responses to DNA damage became a focal point in McGowan's lab at Scripps Research.
Her work on human cells relies on two strains of yeast as model systems, primarily the fission yeast Schizosaccharomyces pombe, which has provided scientists with invaluable leads in their work on human cells. The yeast cell cycle is similar to the human cell cycle, and both yeasts are extremely important: one is similar to the early human cell cycle; the other to the late cell cycle. "They lead us to the right place, but once we get there we find that the landscape looks very different from the yeast configuration," McGowan explains. "So then we have to go to work."
Checking the Checkpoints
This work involves inflicting damage on the DNA of human cells to see how they react. Normal human cells delay at several cell cycle positions when DNA is damaged. These positions are called checkpoints, and these gatekeepers have two critical functions: They stop the cell from dividing so that any damage doesn't become permanent, and they call in the cell's numerous repair processes to take care of the damage.
The checkpoints are enforced by a complex network of kinases, enzymes that alter other proteins by attaching phosphate to them. Checkpoint kinases alter the shape of the proteins and enable them to do the necessary repairs and to halt division. This function points up another characteristic of checkpoints: the fact that they are stalwart guardians. They don't allow the cell to develop further until the damage is repaired. McGowan's lab works primarily on the checkpoints that have to do with G2 and M, sentries in the late cell cycle.
In her lab, McGowan—with human genome sequencing as her map—uses recombinant DNA technology in order to manipulate the proteins of interest, reintroduce them into cells, and watch their activity. "We can tag the proteins to see how they behave after DNA damage," McGowan explains. "These enzymes have very specific activities, so we can run an activity assay on them and separate them using different types of gels to see whose activity went up, whose activity went down, and who got degraded."
Her lab identified two checkpoint kinases in humans that limit progression of the cell cycle when DNA is damaged. One of these kinases, Chk2, short for checkpoint kinase 2, is activated by the protein ATM in response to DNA damage. The gene for ATM is mutated in the cancer-prone disorder ataxia telangiectasia, and cells that lack ATM have diminished responses to DNA damage.
A Cancer-Prone Disorder
McGowan explains that ataxia telangiectasia is a rare disorder affecting children, who at around age seven or eight start to walk unevenly. This symptom is accompanied by black spots in the children's eyes, and the disorder typically progresses to cancer, which eventually kills the patient.
In 1995, scientists in Israel found the gene—the mutation—that causes this disorder, sequenced it, and discovered that it is very similar in sequence to a well-characterized gene in yeast, which is the master regulator of the DNA damage response in that species.
"Since we knew more about the genetic relationships between these proteins in yeast, we could make a very educated guess that there would be a similar relationship in human cells," says McGowan. "We got hold of some of those human cells and tested whether the checkpoint kinases we discovered were downstream of the ataxia telangiectasia protein. And it turned out they are."
McGowan says that one of the reasons this finding is important is that it provides an example of how basic scientific research can lead to understanding of a human disorder. "Because ataxia telangiectasia was a preexisting condition, we knew quite a lot about it, and once we had the yeast connection, we could make an enormous leap in characterizing the human gene and the pathway in general."
Then McGowan returns to Chk2. "The discovery that Chk2 determines whether or not cells die is exciting and important," McGowan says, "because now drugs can be made that target this molecule and that will greatly improve the outcome of radiation therapy."
But, she adds, there are even bigger discoveries still to be made. "We know some of the proteins that Chk2 talks to, but we don't know all of them and don't understand what it does or says to several of the proteins it interacts with. We think we know who most of the players are, but there are enormous gaps in our knowledge about why they're there, what they're doing, and what really happens. By continuing to analyze Chk2 we can make better predictions about how to use it in the clinic, and we can hope to turn up other enzymes that might, for practical reasons, be even better drug targets."
Colleagues, Collaborations, and Camaraderie
The work ahead will be made easier by the scientific camaraderie at Scripps Research, McGowan says, and she singles out one colleague in particular who has played an essential role in her discoveries.
"Paul Russell's lab, right next door, works on the fission yeast model, and my lab relies immensely on that model for defining the parts of the pathways we work on," she says. "So our labs are physically close as well as intellectually close." Russell is a professor in the Department of Molecular Biology.
McGowan adds that findings from Russell's lab are rapidly translated in her lab. "His lab identified an interaction between the checkpoint kinases and a repair protein, and before Paul and his group had taken this finding any further, we were able to identify the same protein in human cells, prove that the same interaction existed, and start working on that almost instantly," she notes. "This synergy, this ease of collaboration, is the name of the game at Scripps."
Send comments to: mikaono[at]scripps.edu
Associate Professor Clare McGowan (left), pictured here with Research Technician Veronique Blais, is fascinated with human cells, particularly how they duplicate. Photo by Kevin Fung.
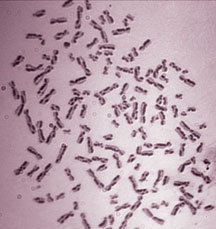
Human chromosomes were stained to distinguish the two pairs of DNA duplex (sister chromatids). Each change in staining pattern, from light to dark or vice versa, represents a point in which exchange between the sisters has been used to repair (or avoid) a problem in one of the strands.
