

CD1 in Prime Time
By Jason Socrates bardi
The present state of human health owes much to our immunological past—millions of years of mammalian evolution through which our highly flexible and effective immune system developed. Without our innate and adaptive immunity, we would not survive long in a world filled with pathogens.
But our immune system is not enough.
In today’s world, our health is helped greatly by the tools we have invented. In the past century, the average life expectancy in the United States has risen from 47.3 years in 1900 to almost 80 years today, due in large part to access to human inventions like modern medical procedures, vaccines, drugs, antibiotics, clean water, proper nutrition, and sanitary living conditions.
However, plenty of old and new infectious diseases still take their toll—bacterial and viral pathogens that cause lethal acute infections. Our ability to continue to improve human health and defeat infectious disease demands that we understand how our immune system interacts with pathogens. The immune system is composed of interacting cells and molecules, but the devil is in the details. What are the players and how are they interacting? In recent years, structural biologists have contributed significantly to our understanding by solving structures that reveal these details and provide a rational basis for the design of new drugs and vaccines
Now, a group of scientists led by Professor Ian Wilson of The Scripps Research Institute is reporting a significant new structure that may greatly improve our understanding of immune recognition. The structure, solved by Scripps Research Associate Dirk Zajonc and Wilson, is CD1a and is one of the key molecules that enable our immune system to sense and respond to common pathogens.
CD1 and its Antigens
CD1a belongs to a family of CD1 receptor proteins (designated CD1a-d) that are present on the surface of two types of immune system cells known as Langerhans and dendritic cells. These are professional antigen presenting cells that play an important role in innate immunity by activating other immune cells in the body during an infection.
Hence, CD1 receptor recognition is vital for defense against common bacteria. It presents T cells in the immune system with specific lipid antigens (fatty molecules that are components of the bacteria). This presentation is what enables the T cells and other components of the immune system to defeat the infection.
A good analogy for this is stock car racing. If a T cell is like a stock car racing through the bloodstream and tracking down bacteria, the professional antigen presenting cells are the pit crew. The car pulls into the pit, and it is “activated” by the crew—presented with new tires and everything it needs to go racing back out on the track. For the pit crew to work, of course, it needs the right kind of tires.
The tire is analogous to the lipid molecule. Antigen presenting cells use CD1 molecules loaded with a lipid antigen to activate T cells with receptors specific for that antigen. Once an inactive T cell binds to CD1, it will become activated and unleash a torrent of action aimed at clearing the infectious agent.
These T cells expand in number and, depending on whether they are killer or helper T cells, begin killing bacteria or secreting large amounts of inflammatory immune proteins. This in turn activates other parts of the immune system—for instance, inducing specific B cells to unload bursts of soluble antibodies into the bloodstream, which target the pathogens for destruction.
Scientists would like to know the full range of lipids that CD1 binds and presents to other immune cells and the structure of CD1 loaded with antigen bound to a T cell receptor. Not surprisingly, waves of immunologists have studied CD1a and other CD1 proteins, and to date studies have examined these proteins in every sort of mammal from mice to humans. However, these molecules have only begun revealing their secrets.
About 10 years ago, a group of scientists at Harvard Medical School identified the first antigen bound by CD1, and since then several others have been identified. These include both natural human lipids and those from the cell walls of common bacteria, such as Mycobacterium tuberculosis and Nisseria gonorrheae. Even so, scientists have not been able to identify all of the antigens bound by CD1, and they had a hard time until recently elucidating the structural details of this important family of proteins bound to a bacterial antigen.
Structures Abound
Two years ago, Zajonc, Wilson and their colleagues solved a structure of CD1a complexed with the molecule sphingolipid—a human molecule. But the scientists wanted to solve a structure of CD1a with a microbial antigen attached to get a glimpse of how CD1 activates a T cell.
Last year, Branch Moody, a collaborator of Wilson and Zajonc’s at Harvard Medical School, identified a new class of bacterial antigens presented by CD1a molecules: a lipopeptide, which is part lipid and part amino acid.
These lipopeptides, called dideoxymycobactins, are precursors to the iron binding molecules called siderophores that are part of an iron scavenging system that many bacteria have to steal iron from the host. Iron is precious to bacteria because they use it to ferry electrons around during metabolic processes, and bacteria release siderophores into human host cells upon infection.
In an article that appears as the cover story of last month’s issue of the journal Immunity, Zajonc, Wilson, Moody, and their colleagues are reporting the crystal structure of CD1a bound to the lipopeptide dideoxymycobactins from Mycobacterium tuberculosis, the infectious agent that causes TB. This is the first such structure of a CD1 molecule with a microbial antigen bound. It is also the first time the structure of a lipopeptide that is presented to the immune system has been solved—potentially an important general class of molecules presented by CD1 and, as such, interesting vaccine candidates.
The structure shows how single-chain lipopeptide can maneuver into the CD1a binding pocket. The binding groove of CD1 is a narrow and deep hydrophobic trough, into which the hydrophobic lipid molecules readily sink. The lipopeptide is unusual in that it has only one single lipid tail (most of the lipids that are known to bind to CD1 have two). To the molecule’s single alkyl chain, a highly modified peptide is attached.
The structure shows that the one lipid chain sticks into one end of the hydrophobic pocket of the CD1a protein. And where a two-lipid-chain molecule might stick its other lipid, the lipoprotein inserts part of its peptide chain—that includes a curious amino acid made of lysine that has been cyclized. This exposes the rest of the peptide for recognition by T cells.
After solving the structure, Zajonc and Wilson also modeled a T cell receptor docked onto the CD1 protein, based on a T cell receptor structure that was solved in another study. The Holy Grail for the field, says Zajonc, is to solve the structure of CD1 with an actual T cell receptor docked to it that would elucidate how the immune system recognizes lipid molecules and could be used to provide new therapies.. This has been elusive for a number of reasons, he adds, not the least of which is the difficulty in producing enough soluble T cell receptors for structural studies.
The work is ongoing.
To read the article, “Molecular Mechanism of Lipopeptide Presentation by CD1a” by Dirk M. Zajonc, M.D. Max Crispin, Thomas A. Bowden, David C. Young, Tan-Yun Cheng, Jingdan Hu, Catherine E. Costello, Pauline M. Rudd, Raymond A. Dwek, Marvin J. Miller, Michael B. Brenner, D. Branch Moody and Ian A. Wilson, see the February, 2005 issue of the journal Immunity (209-219) or go to http://dx.doi.org/10.1016/j.immuni.2004.12.009.
Send comments to: jasonb@scripps.edu

Professor Ian Wilson (left) and Research Associate Dirk Zajonc solved the structure of CD1a, a key molecule that enables our immune system to sense and respond to common pathogens. Photo by Jason S. Bardi.
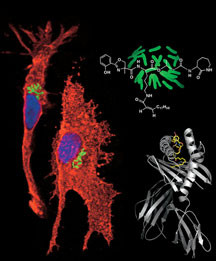
How peptide sequences can be presented to and recognized by T cells. Click to Enlarge.
