

From Molecules to Medicine:
Understanding Gap Junction Channels
By Jason Socrates Bardi
"The third [logical precept is] to conduct my thoughts in such order that, by commencing with objects the simplest and easiest to know, I might ascend by little and little, and, as it were, step by step, to the knowledge of the more complex."
—Rene Descartes, Discourse on the Method of Rightly Conducting the Reason and Seeking Truth in the Sciences, 1637
In an interview late one recent autumn afternoon in his office at The Scripps Research Institute, Cell Biology Professor Mark Yeager was discussing gap junction channels with a visitor—something that he has done many times in the almost two decades that he has been studying these proteins. As he had done many times before, Yeager was listing the nuanced ways in which gap junction channels contribute to health and disease and describing why they are so interesting.
So why?
To begin with, gap junction channels have particularly interesting properties, Yeager says. They are assemblies of proteins that come together to form pores between adjacent cells, and in so doing, coordinate the communication within tissues throughout the body. They coordinate communication between cells in the heart, the skin, the liver, and they even coordinate the rhythmic contractions of the uterus during birth.
There's more. Like all forms of communication, the dialog between cells is not always accurate. The gap junction channels sometimes do not function properly because the pores may be closed or the protein subunits may not even assemble properly within cell membranes. Such "molecular pathology" can cause a variety of health problems, such as heart attacks, deafness, and neurological disorders. It is understanding the relationship between the molecular structure of gap junction channels and their relevance to medicine and diseases that drives Yeager's research.
Recently, Yeager and his collaborators, Sarel Fleishman and Nir Ben-Tal at the Tel-Aviv University in Israel and Vinzenz Unger at Yale University, combined the results of electron crystallography and computational modeling to derive a hypothetical molecular structure for gap junction channels (see figure). This model served as a template on which to map a number of known amino acid mutations in the proteins that form gap junction channels, which result in a variety of diseases. Their results were published in the September 24 issue of the journal Molecular Cell.
A surprising result of their analysis is that almost all of the mutations occur at the interfaces between a-helices—long, compact spirals that traverse the membrane. (a-helices are a fundamental motif of protein structure that was discovered by Linus Pauling, for which he was awarded the Nobel Prize in 1954.) The fact that the mutations occur in these a-helices suggests clues about the underlying molecular basis of disparate diseases related to gap junction channels.
"Precise packing of the a-helices is critical for the normal assembly and function of the channels," says Yeager.
Junction, Junction, What's Your Function?
The easiest way to understand gap junctions is to look at them in the context of what they do in the tissue that is closest to Yeager's heart—in fact, the heart itself.
Anyone who has ever ridden a bike up the long, steep Torrey Pines Mesa, upon which Scripps Research's La Jolla campus is situated, can hear that thump-thumping in their ears—perhaps to the exclusion of any other sound, depending on what sort of shape the rider is in.
However consumed a rider is by his beating heart, he may not be aware that each heartbeat is actually a coordinated contraction of hundreds of thousands of cardiac muscle cells throughout the heart, which depends on proper functioning of the gap junction channels.
The heartbeat starts with a small cluster of so-called pacemaker cells in a region of the heart known as the sinoatrial node. These cells beat spontaneously, generating an electrical current or "action potential," which spreads throughout the heart via a specialized wiring system of cells connected by gap junctions. Likewise, the current then spreads between all the muscle cells via gap junction channels.
This process of current flow to coordinate the contraction of cardiac myocytes takes place in only about 100 milliseconds, and this process is repeated more than 100,000 times each day throughout your life. If heart muscle cells were not able to perform this continuous and highly coordinated contraction, the heart would not be able to effectively pump blood to and from the lungs and into peripheral tissues. Aberrant beating patterns, known as heart arrhythmias, occur due to abnormal patterns of current flow through the gap junction channels. Indeed, about one person dies each minute due to sudden cardiac death.
Structure of the Cell Dialogue
Yeager's early experiments on gap junction channels involved the use of antibodies targeted to specific sites in the protein to identify those amino acid residues that form loops outside of the cell membranes. But these sorts of experiments only gave a crude picture of the channel.
Nowadays scientists have a much more detailed understanding of the channel, in part due to an electron microscope structure of the heart gap junction channel that Yeager and his colleagues solved and published in a cover article in the journal Science in 1999.
Electron microscopy (or EM) has been around since the first electron microscope was built by Ernst Ruska in 1933 (for which he received the Nobel Prize in 1986 at age 80). These microscopes use magnetic lenses to bend a beam of electrons to image tiny objects, similar to the bending of light by glass lenses in a light microscope. EM looks at a range of magnifications, from no more than an ordinary light microscope that magnifies up to 60 times to those that magnify up to 1,000,000 times.
Scripps Research is one of the few scientific centers in the world with an integrated program in electron microscopy of biological complexes and macromolecular machines, but even with so many resources, it is not always a simple matter to solve a complicated structure like a gap junction channel.
To address this problem, Yeager's group exploited the natural tendency of gap junction channels to aggregate into plaques within cell membranes. The real breakthrough that allowed the 1999 structure to be solved was the development of the ability to make well ordered two-dimensional crystals of recombinant gap junction channels. Images of crystals at different tilt angles were recorded in the electron microscope, and the crystallographic data could be merged to yield a 3D map at fairly high resolution—about 6 Ångstoms currently. The 2D crystals are a far cry from the almost perfect arrays of ordered protein molecules in X-ray crystallography that yields structures at atomic resolution. Nevertheless, they are the only alternative for the biochemically complex gap junction, and much was learned from the EM structure.
A single heart gap junction channel is formed by two "hemichannels," one in each of the two adjoining cells. Each hemichannel, which is called a "connexon" is composed of six protein subunits (each individual subunit is called a "connexin") in the membrane of one of the adjoining cells. Each connexin subunit is formed by 4 a-helical rods that traverse the membrane.
A single connexon hemichannel, then, has 24 of these a-helices spanning the cell's membrane, and a complete gap junction channel forms between two adjacent cells by the end-to-end docking of these connexon hemichannels. So the entire assembly has a remarkable structure of 48 a-helices. Once two hemichannels dock end-to-end, the fully functional gap junction channel they form has a central pore, sort of like a molecular conduit that allows small metabolites and ions to travel from cell to cell.
All in the Family of Gap Junctions
Scans of the human genome have yielded dozens of different proteins that have been grouped together into the family of gap junction proteins, called connexins. The assumption, which is usually correct, is that proteins with similar enough sequences will have similar structures, and will then engage in similar molecular or physiological functions.
Although not at the resolution achieved in X-ray crystallography, Yeager says, "The current EM map defines the position and orientation of the transmembrane a-helices."
In their paper in the journal Molecular Cell, Yeager's collaborators Nir Ben-Tal and Sarel J. Fleishman used the EM map of connexin-43 as a template for building a general model of the gap junction channel by computational analysis of all the known connexin amino acid sequences. The known mutations in connexins could then be mapped onto this model.
Over the past few decades, a number of pathological conditions in humans and in mouse models of disease have been attributed to mutations in connexins. For instance, more than two dozen different mutations in connexin-26 are associated with the most common form of nonsyndromic deafness—a congenital form of deafness in which the children suffer no additional complications, such as blindness or mental retardation.
Several mutations in connexin-32 are associated with Charcot-Marie-Tooth disease, which is one of the most common inherited neurological disorders in the United States, according to the National Institute of Neurological Disorders and Stroke. The disease, named after its nineteenth-century co-discoverers, the physicians Jean-Martin Charcot, Pierre Marie, and Howard Henry Tooth, afflicts approximately one in 2,500 Americans with peripheral nerve disorders. These disorders have a range of effects on patients, including movement problems, motor skill difficulties, and muscular atrophy.
In the gap junction model, almost all of the connexin-26 and -32 mutations were located at the interfaces between the a-helices, which provides an intriguing molecular explanation for those diseases. The mutations are occurring primarily in regions of the gap junction where the amino acids are tightly packed and, thus, less able to tolerate variations in the amino acid sequence.
Still a Long Road Ahead
Yeager is enthusiastic about the results, he says, because this is the first time he and his colleagues have had a sufficiently detailed model for the gap junction channel so that it could be related to human disease. However, he warns, "Understanding a molecular structure and the mechanism [of a disease] doesn't immediately provide a treatment."
Yeager points to the example of sickle-cell anemia, which is the most common inherited blood disorder in the United States. The disease is caused by defects in the oxygen-transporting protein hemoglobin that causes these proteins to stick together and twist the red blood cells into crescent sickle shapes. The sickled cells do not have the pliability to move through the microvasculature, and the sludging of the cells causes intense pain and tissue damage.
Scientists have known for decades that the substitution of the hydrophobic amino acid valine with the charged amino acid glutamate causes the hemoglobin molecules to stick together and form fibers. And Max Perutz received the Nobel Prize in 1962 for solving the atomic structure of hemoglobin. In spite of these discoveries, there is still no breakthrough for curing the disease. Doctors today treat sickle cell patients using largely the same procedures they did when Yeager was in medical school—IV hydration, transfusions and painkillers.
"Still," Yeager says, "we think it's essential to understand the molecular basis of disease to even contemplate a treatment."
Send comments to: jasonb@scripps.edu

"We think it's essential to understand the molecular basis of disease to even contemplate a treatment," says Professor Mark Yeager.
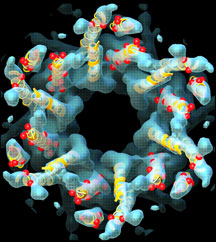
Intercellular gap junction channel. Click to enlarge and read caption.
Links:
