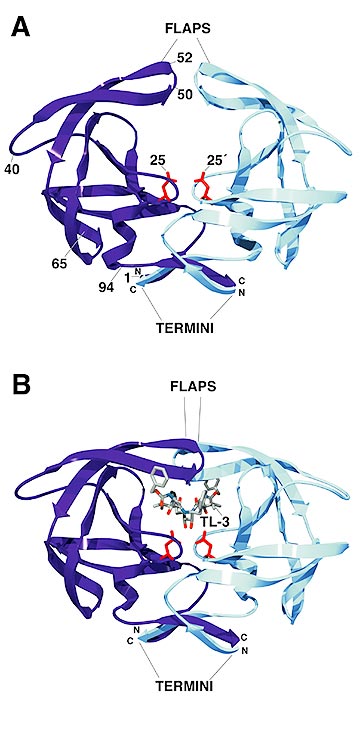
The homodimeric aspartyl protease undergoes dramatic conformational change in its so-called "flaps" region upon binding of inhibitors. The Scripps Research Institute protease inhibitor, TL-3, is no exception. Here, we can see the protease in its unbound, apo-form in (A), and in the crystal structure solved by our NCI/FCRF collaborators Alex Wlodawer and Alla Gustchina, of the complex with TL-3. These structures can be found in the Protein Data Bank under the PDB codes 1HHP and 3TLH, respectively. Note the location of the catalytic aspartic acids in the centre of the active site (residues 25 and 25').